Sequential Simplex Method: A Practical Guide for Optimizing Multi-Factor Experiments in Biomedical Research
This comprehensive guide explores the sequential simplex method, a powerful model-agnostic optimization technique ideal for researchers and drug development professionals navigating complex experimental spaces with multiple interacting factors.
Sequential Simplex Method: A Practical Guide for Optimizing Multi-Factor Experiments in Biomedical Research
Abstract
This comprehensive guide explores the sequential simplex method, a powerful model-agnostic optimization technique ideal for researchers and drug development professionals navigating complex experimental spaces with multiple interacting factors. The article covers foundational principles, from the algorithm's origins in George Dantzig's work to its geometric interpretation of navigating response surfaces. It provides detailed methodological guidance on implementing both basic and modified simplex procedures, illustrated with real-world applications from analytical chemistry and biopharmaceutical production. The guide also addresses critical troubleshooting considerations for managing experimental noise and step size selection, while offering comparative analysis against alternative optimization approaches like Evolutionary Operation and Response Surface Methodology. Designed for practical implementation, this resource enables scientists to efficiently optimize processes ranging from analytical method development to recombinant protein production and drug formulation.
The Geometry of Optimization: Understanding Sequential Simplex Fundamentals
Frequently Asked Questions (FAQs)
FAQ 1: What is the core connection between George Dantzig's Linear Programming and modern experimental optimization?
George Dantzig's Simplex Algorithm, developed in 1947, provides the mathematical foundation for optimizing a linear objective function subject to linear constraints [1] [2]. Modern experimental optimization builds upon this by applying the same core principle—systematically moving toward an optimum—to the real-world process of experimentation. While Dantzig optimized mathematical models, researchers now use methods like the sequential simplex to optimize experimental factors themselves, efficiently navigating multi-factor spaces to find the best combinations for desired outcomes [3].
FAQ 2: Why should I move beyond one-factor-at-a-time (OFAT) experiments?
One-factor-at-a-time (OFAT) experimentation is inefficient and can lead to misleading conclusions [4]. Crucially, OFAT cannot detect interactions between factors—situations where the effect of one factor depends on the level of another [3] [4]. For example, in the corn cultivation case, the effect of fertilizer was different for basic corn versus MegaCorn Pro [4]. Multi-factor factorial designs, in contrast, allow you to efficiently explore many variables simultaneously and discover these critical interactions, leading to more robust and optimal results [3].
FAQ 3: How does the Sequential Simplex Method improve the efficiency of my experiments?
The Sequential Simplex Method is an iterative, "hill-climbing" optimization procedure [5]. It improves efficiency by using the results from one experimental run to determine the most promising conditions for the next run. This creates a direct, adaptive path towards optimal factor settings, avoiding the wasted effort of testing non-informative conditions. It systematically moves from an initial basic feasible solution to adjacent solutions with better objective function values until the optimum is found [1] [5].
FAQ 4: What are the common signs that my experimental optimization is failing to converge?
- Cycling: The simplex repeats a sequence of points without progressing to a new, better solution [5].
- Oscillation: The results fluctuate between similar values without stabilizing.
- Lack of Improvement: The objective function (e.g., yield, purity) shows no significant improvement over multiple iterations.
- Violation of Constraints: Proposed experimental conditions are infeasible or violate safety or practical limits.
FAQ 5: How do I validate the optimal conditions found by a sequential simplex procedure?
- Replication: Perform multiple experimental runs at the purported optimal conditions to confirm reproducibility and estimate variability.
- Confirmation Run: Execute a final, controlled experiment using the optimal factor settings to verify that the predicted performance is achieved.
- Model Validation: If a predictive model was used, validate it with a new set of data not used in the optimization process [6].
Troubleshooting Guides
Problem 1: The Simplex Algorithm Fails to Find an Improved Solution
Symptoms:
- The algorithm cycles through the same solutions.
- The objective function value does not improve after an iteration.
| Possible Cause | Diagnostic Steps | Solution |
|---|---|---|
| Degeneracy [5] | Check if a basic variable has a value of zero in the feasible solution. | Apply an anti-cycling rule (e.g., Bland's rule) to perturb the solution slightly and break the cycle [1]. |
| Incorrect Pivot Selection | Verify the calculations for the reduced cost coefficients (entering variable) and the minimum ratio test (leaving variable) [1] [7]. | Recalculate the simplex tableau. Ensure the most negative reduced cost is chosen for maximization and the minimum ratio is correctly identified. |
| Local Optimum | The solution may be a local, not global, optimum. This is less common in pure LP but possible in non-linear response surfaces. | Restart the algorithm from a different initial basic feasible solution to explore other regions of the feasible space. |
Problem 2: Poor Optimization Performance in Multi-Factor Experiments
Symptoms:
- High variability in responses.
- Inability to identify significant factors.
- The predicted optimum does not perform well in validation.
| Possible Cause | Diagnostic Steps | Solution |
|---|---|---|
| Ignored Factor Interactions [4] | Analyze the data for two-factor interactions. A significant interaction means the effect of one factor depends on the level of another. | Shift from a OFAT design to a full or fractional factorial design that can estimate interactions [3] [4]. |
| Uncontrolled Noise | Review the experimental setup for sources of uncontrolled variability (e.g., environmental conditions, operator differences). | Introduce blocking into the experimental design to account for known sources of noise and reduce background variation [4]. |
| Incorrect Region of Experimentation | The initial range of factors being tested may be far from the true optimum. | Perform a screening design first to identify important factors, then use a response surface method (like simplex) to hone in on the optimum. |
Problem 3: Infeasible or Unrealistic Solutions from the Model
Symptoms:
- The algorithm indicates no feasible solution exists.
- The proposed optimal factor settings are impractical or dangerous to implement.
| Possible Cause | Diagnostic Steps | Solution |
|---|---|---|
| Over-constrained System | Check all constraints for consistency. Even a single contradictory constraint can make the entire problem infeasible [1]. | Re-examine the necessity and values of each constraint. Relax constraints if possible and scientifically justified. |
| Incorrectly Formulated Constraints | Verify that all variable bounds (e.g., temperature > 0, concentration ≤ 100%) are correctly specified. | Reformulate the constraints and variable bounds to accurately reflect the physical and practical limits of the experiment [1]. |
| Model does not reflect reality | The linear or simplified model may be inadequate for the complex system under study. | Consider using a more complex, non-linear model or incorporating domain expertise to refine the experimental setup and constraints. |
Experimental Protocols & Methodologies
Protocol 1: Setting Up a Initial Simplex for a New Experiment
Purpose: To create a starting simplex for optimizing multiple factors.
- Select Factors: Choose the
kcontinuous factors you wish to optimize. - Define Range: Establish a feasible operating range for each factor.
- Create Initial Simplex: Generate
k+1experimental runs that form the initial simplex in thek-dimensional factor space. For example, for 2 factors, a triangle is formed. - Run Experiments: Execute the
k+1experiments in a randomized order to minimize the effect of lurking variables. - Measure Response: Record the objective function value (e.g., yield, efficiency) for each run.
Protocol 2: Executing a Sequential Simplex Iteration
Purpose: To move the simplex towards a more optimal region.
- Evaluate Vertices: Rank the responses of all current simplex vertices.
- Identify Worst: Identify the vertex with the worst response.
- Reflect: Reflect the worst point through the centroid of the opposite face to generate a new candidate point.
- Test New Point: Run the experiment at the new candidate point.
- Compare & Decide:
- If the new point is better than the worst, replace the worst point with it.
- If the new point is the best so far, consider a further expansion step.
- If the new point is worse than the second-worst, consider a contraction step.
- Check Termination: Continue iterating until the simplex converges around an optimum or a predefined number of runs is completed.
The Scientist's Toolkit: Research Reagent Solutions
| Item | Function in Experimental Optimization |
|---|---|
| Slack Variables [1] [7] | Convert inequality constraints (e.g., "resource use ≤ budget") into equalities, making them usable in the simplex algorithm. They represent "unused" resources. |
| Simplex Tableau [1] [7] | A tabular format used to perform the algebraic manipulations of the simplex algorithm efficiently. It organizes the coefficients of the objective function and constraints. |
| Factorial Design [3] [4] | An experimental design that studies the effects of two or more factors, each with multiple levels. It is used to screen for important factors and estimate interactions before detailed optimization. |
| Fractional Factorial Design [3] | A more efficient version of a full factorial design that tests only a carefully chosen subset of all possible combinations. It is used when the number of factors is large, trading some interaction detail for speed and lower cost. |
| Objective Function | The single quantitative measure (e.g., process yield, product purity, cost) that the experiment is designed to optimize. |
| 2-Iodoadenosine | 2-Iodoadenosine, CAS:35109-88-7, MF:C10H12IN5O4, MW:393.14 g/mol |
| Fenticonazole | Fenticonazole | Antifungal Agent for Research (RUO) |
Workflow and Relationship Diagrams
Sequential Simplex Optimization Flow
Multi-Factor Experiment Design Logic
Troubleshooting Guides and FAQs
Frequently Asked Questions
Q1: What does it mean if my simplex is oscillating between the same few vertices? This typically indicates that the simplex is circling a potential optimum. According to Rule 2 of the sequential simplex method, when a new vertex yields the worst result, you should reject the vertex with the second-worst response instead to change the direction of progression [8] [9]. This helps the simplex navigate more effectively in the region of the optimum.
Q2: How do I handle a situation where the new experimental conditions fall outside feasible boundaries? Rule 4 of the method provides a solution: assign an artificially worst response to any vertex that falls outside the experimental boundaries [8]. This forces the simplex to reject this point and move back into the feasible experimental domain on the next iteration.
Q3: My optimization progress has become very slow. How can I improve convergence? The basic simplex method maintains a fixed size, which can lead to slow progress. Consider switching to the modified simplex method by Nelder and Mead, which allows for expansion and contraction steps [8] [9]. An expansion move is performed if the reflected vertex yields a much better response, rapidly accelerating progress toward the optimum.
Q4: A single vertex remains in many successive simplexes. What does this signify? When a point is retained in f+1 successive simplexes (e.g., 4 simplexes for 3 factors), it suggests a possible optimum. Apply Rule 3: re-run the experiment at this vertex [8]. If it confirms the best response, it is likely the optimum. If not, the simplex may be stuck at a false optimum, and a restart with a different initial simplex may be necessary.
Common Error Scenarios and Solutions
| Error Scenario | Possible Cause | Recommended Solution |
|---|---|---|
| Simplex Oscillation | Simplex circling near optimum [8]. | Apply Rule 2: Reflect second-worst vertex. |
| Slow Convergence | Fixed simplex size is too small for the region [9]. | Use modified method with expansion/contraction. |
| Boundary Violation | New vertex coordinates are outside feasible domain [8]. | Apply Rule 4: Assign artifical worst response. |
| False Optimum | Vertex retained but not truly optimal [8]. | Apply Rule 3: Re-test vertex; consider re-start. |
| High Dimensionality | Number of factors (f) is large, increasing complexity [8]. | Eliminate linear parameters first to reduce dimensions [9]. |
Experimental Protocol: Sequential Simplex Method
Objective
To systematically optimize multiple experimental factors by navigating the multi-dimensional factor space using a sequential simplex algorithm to find the combination that yields the optimal response.
Methodology
1. Initial Simplex Construction
- For an experiment with
ffactors, construct an initial simplex withf+1vertices [8] [9]. - Each vertex represents a unique set of experimental conditions (e.g., specific concentrations, temperatures).
- The size of the initial simplex should be chosen arbitrarily based on the experimental domain [8].
2. Experiment Execution and Evaluation
- Run the experiment at each vertex of the current simplex.
- Measure the response for each vertex (e.g., yield, purity, activity).
3. Vertex Ranking and Movement
- Rank all vertices from Best (B) to Worst (W), with N representing the next-to-best [8] [9].
- Calculate the Centroid: Compute the centroid (P) of all vertices except the worst (W). For
ffactors, the centroid's coordinates are the average of the coordinates of the retained vertices [8]. - Reflect the Worst Vertex: Generate a new vertex (R) by reflecting the worst vertex through the centroid. The vector formula is: R = 2P - W [8].
4. Decision Logic for Simplex Progression The following workflow outlines the core logic for moving from one simplex to the next, including the reflection, expansion, and contraction operations.
5. Iteration and Termination
- Repeat steps 2-4, using the decision logic above, until the simplex converges on the optimum region.
- Convergence is typically signaled when the simplex tightly circles a point with little improvement in response over multiple iterations [8] [9].
- The vertex with the best observed response is reported as the optimal set of conditions.
The Scientist's Toolkit: Research Reagent Solutions
| Item or Concept | Function in Sequential Simplex Optimization |
|---|---|
| Initial Simplex | The starting geometric figure in factor space; its size and location set the initial scope of the investigation [8]. |
| Factor Space | The n-dimensional coordinate system defined by the n factors being optimized; the "landscape" being navigated [8]. |
| Response Surface | The often-unknown multidimensional surface representing the outcome (response) for every combination of factors; the simplex method navigates this surface without requiring its explicit mapping [9]. |
| Centroid (P) | The geometric center of the face opposite the worst vertex (W); the pivot point for reflection, expansion, and contraction moves [8]. |
| Reflection | The core operation that projects the worst vertex through the centroid to explore a new, potentially better region of the factor space [8] [9]. |
| Expansion | (Modified Method) An operation following a successful reflection, which extends the simplex further in that direction to accelerate progress if the reflection was much better [9]. |
| Contraction | (Modified Method) An operation used when a reflection is poor, which shrinks the simplex to hone in on a promising area [9]. |
| Linear Optimization Oracle | In advanced geometric methods, an efficient algorithm used to optimize a linear function over the feasible set polytope, aiding in decomposition for neural combinatorial optimization [10]. |
| Perospirone | Perospirone, CAS:150915-41-6, MF:C23H30N4O2S, MW:426.6 g/mol |
| Fmoc-Glu(OAll)-OH | Fmoc-Glu(OAll)-OH, CAS:133464-46-7, MF:C23H23NO6, MW:409.4 g/mol |
Frequently Asked Questions (FAQs)
Q1: What is the fundamental geometric concept behind the Basic Simplex Algorithm?
The Basic Simplex Algorithm uses a regular geometric figure called a simplex to navigate the experimental domain [11] [9]. For an optimization involving k factors or variables, the simplex is defined by k+1 vertices [12] [9]. In two dimensions, this simplex is a triangle; in three dimensions, it is a tetrahedron; and for higher dimensions, it is a hyperpolyhedron or simplicial cone [1] [11] [12]. The algorithm proceeds by moving this fixed-size figure through the experimental space toward the optimal region.
Q2: What does "fixed-size" mean in the context of the Basic Simplex method?
"Fixed-size" means that the geometric figure (the simplex) does not change in size during the optimization process [12] [9]. The initial simplex, defined by the researcher, remains a regular figure with constant edge lengths as it is reflected across its faces. This is a key difference from the Modified Simplex method, where the simplex can expand or contract [12] [9].
Q3: What are the primary moves or operations in the Basic Simplex Algorithm?
The core operation in the Basic Simplex method is reflection [9]. At each step, the vertex yielding the worst response is identified and rejected. This worst vertex is then reflected through the opposite face (or line, in two dimensions) of the simplex to generate a new vertex. The new vertex and the remaining vertices from the previous simplex form the new simplex [9]. This reflection process is repeated sequentially.
Q4: What rules govern the movement of the simplex?
The algorithm is governed by two main rules [9]:
- Rule 1: The new simplex is formed by reflecting the worst vertex through the face defined by the remaining vertices. This is the standard move.
- Rule 2: If the newly reflected vertex is again the worst in the new simplex, the vertex with the second-worst response is reflected instead. This rule prevents the simplex from oscillating between two points and helps change direction, particularly when near the optimum.
Q5: What is a critical step for the researcher when using the Basic Simplex method?
Choosing the size of the initial simplex is a crucial and often difficult step [12]. Because the simplex size remains fixed, an initial simplex that is too large may miss fine details of the response surface, while one that is too small will make the progression toward the optimum very slow. This decision often relies on the researcher's experience and prior knowledge of the system being studied [12].
Troubleshooting Guide
| Problem | Possible Cause | Solution |
|---|---|---|
| Slow Convergence | The initial simplex size is too small. | Use prior knowledge of the system to choose a larger, more appropriate initial simplex size [12]. |
| Oscillation (Simplex moves back and forth between two points) | The simplex is reflecting the worst vertex back to its original position (a violation of Rule 1) [9]. | Apply Rule 2: Identify and reflect the vertex with the second-worst response instead of the worst one to change direction [9]. |
| Missing the Optimal Region | The initial simplex size is too large, making it difficult to locate the precise optimum [12]. | Restart the optimization with a smaller initial simplex focused on the most promising area found in the initial broad search. |
| Circling Around a Point | The simplex is likely in the vicinity of the optimum. Subsequent moves keep one vertex (the best one) constant [9]. | The retained vertex is likely near the optimum. Terminate the procedure or use the final simplex to define a new, smaller simplex for a more precise location. |
Research Reagent Solutions
The following table outlines the essential conceptual "reagents" or components needed to set up a Basic Simplex experiment.
| Item | Function in the Experiment |
|---|---|
| Initial Simplex | The regular geometric starting figure (e.g., a triangle for 2 variables) defined by k+1 experimental points. It sets the scope and direction of the optimization [12] [9]. |
Objective Function (f(x)) |
The measurable response (e.g., yield, sensitivity, signal-to-noise ratio) that the algorithm seeks to minimize or maximize [11]. |
Experimental Variables (x1, x2,... xk) |
The independent factors (e.g., temperature, pH, concentration) that are adjusted to optimize the objective function [12]. |
| Reflection Operation | The computational procedure that generates a new candidate vertex by mirroring the worst vertex, enabling the simplex to move through the experimental domain [9]. |
| Stopping Criterion | A pre-defined rule (e.g., no significant improvement after several steps, or circling behavior) to halt the optimization process [9]. |
Basic Simplex Movement Protocol
The diagram below visualizes the logic and rules governing the movement of a fixed-size simplex.
The table below summarizes the key characteristics of the movements in the Basic Simplex Algorithm.
| Aspect | Description in Basic Simplex |
|---|---|
| Figure Dimensionality | k dimensions, defined by k+1 vertices, where k is the number of experimental variables [12] [9]. |
| Primary Move | Reflection [9]. |
| Figure Size | Fixed throughout the procedure [12] [9]. |
| Rules for Direction Change | Rule 2 (Reflect the second-worst vertex) is applied when reflection of the worst vertex fails [9]. |
| Typical Termination Signal | The simplex begins to circle around a single, retained vertex [9]. |
Frequently Asked Questions
Q1: What is the fundamental difference between the Nelder-Mead method and the traditional simplex algorithm for linear programming?
The Nelder-Mead method is a direct search heuristic for nonlinear optimization problems where derivatives may not be known, and it uses a geometric simplex of n+1 points in n dimensions [13]. In contrast, the traditional simplex algorithm developed by Dantzig is designed exclusively for linear programming problems and operates by moving along the edges of the feasible region defined by linear constraints to find the optimal solution [1]. The two methods are distinct and should not be confused.
Q2: During an iteration, my reflected point is better than the current best point. What operation does the algorithm perform next, and what is the purpose?
This scenario triggers an Expansion operation [13] [14]. The algorithm has found a highly promising direction, so it tests an expansion point further out along the reflection direction. The purpose is to accelerate progress downhill by taking a larger step in this promising direction. If the expansion point is better than the reflection point, it is accepted; otherwise, the reflected point is accepted [15].
Q3: What does the algorithm do when the reflection point is worse than the worst point in the simplex?
When the reflection point is worse than the worst point, the algorithm attempts a Contraction Inside operation [13] [14]. It tests a point between the worst point and the centroid. If this inside contraction point is better than the worst point, it is accepted. If not, the algorithm performs a Shrink operation, moving all points (except the best) towards the best point to refine the search area [15].
Q4: How should I initialize the simplex, and what are common termination criteria?
A common initialization strategy, used in implementations like MATLAB's fminsearch, starts from a user-given point xâ‚€. The other n vertices are set to xâ‚€ + Ï„áµ¢eáµ¢, where eáµ¢ is a unit vector, and Ï„áµ¢ is a small step (e.g., 0.05 if the component is non-zero, 0.00025 if it is zero) [16]. Termination is often based on the simplex becoming small enough or the function values at the vertices becoming sufficiently close [14] [16].
Troubleshooting Common Experimental Issues
Problem 1: The optimization converges to a non-stationary point or gets stuck.
- Potential Cause: The Nelder-Mead method is a heuristic and can converge to points that are not true minima, especially on problems that do not meet stronger conditions required for modern methods [13].
- Solution: Restart the algorithm from different initial points and compare the results. Empirical evidence suggests that multiple shorter runs can yield better overall performance than a single long run [15].
Problem 2: The algorithm is making very slow progress in later iterations.
- Potential Cause: The simplex has shrunk very small and is oozing down a shallow valley [13], or it is trying to "pass through the eye of a needle" [13].
- Solution: This is part of the expected behavior as the algorithm contracts around a suspected minimum. Check the termination criteria; if the standard deviation of the function values or the size of the simplex is below your tolerance, the algorithm can be stopped [16].
Problem 3: The algorithm fails to improve the worst point after reflection, expansion, and contraction attempts.
- Potential Cause: This is a known rare situation where the simplex is on the face of a "valley" or the problem is highly irregular [15].
- Solution: The algorithm handles this by default with a Shrink operation. Shrinking the simplex towards the best point helps the algorithm to reorient itself and can be crucial for escaping these stuck configurations [13] [15].
Experimental Protocol: Implementing the Nelder-Mead Algorithm
This protocol outlines the core iterative procedure of the Nelder-Mead method, focusing on the expansion and contraction operations.
1. Initialization
- Define the objective function
f(x)to be minimized and the number of dimensionsn. - Construct an initial simplex of
n+1vertices. For a given starting pointx₀, a common approach is to create the other vertices asx₀ + τᵢeᵢ, whereeᵢare unit vectors andτᵢare small step sizes [16].
2. Algorithm Iteration Repeat the following steps until a termination criterion is met:
- Step 1: Ordering. Evaluate
f(x)at each vertex and order the points so thatf(xâ‚) ≤ f(xâ‚‚) ≤ ... ≤ f(xₙ₊â‚). Identify the best (xâ‚), worst (xₙ₊â‚), and second-worst (xâ‚™) points [13]. - Step 2: Calculate Centroid. Calculate the centroid
xâ‚’of thenbest points (excludingxₙ₊â‚) [13]. - Step 3: Reflection.
- Step 4: Expansion.
- Step 5: Contraction.
- This step is reached if
f(x_r) ≥ f(xₙ). - Outside Contraction: If
f(x_r) < f(xₙ₊â‚), compute the outside contraction point:x_c = xâ‚’ + Ï(x_r - xâ‚’)where0 < Ï â‰¤ 0.5(typicallyÏ=0.5). Iff(x_c) ≤ f(x_r), acceptx_c; otherwise, go to Step 6: Shrink [13]. - Inside Contraction: If
f(x_r) ≥ f(xₙ₊â‚), compute the inside contraction point:x_c = xâ‚’ + Ï(xₙ₊₠- xâ‚’). Iff(x_c) < f(xₙ₊â‚), acceptx_c; otherwise, go to Step 6: Shrink [13].
- This step is reached if
- Step 6: Shrink. If no improvement was found via reflection or contraction, perform a shrink. Replace all points except the best (
xâ‚) withx_i = xâ‚ + σ(x_i - xâ‚)for alli, where0 < σ < 1is the shrink coefficient (typicallyσ=0.5) [13]. Begin the next iteration with the new simplex.
3. Termination
- Common termination criteria include: the function values at all vertices are within a specified tolerance (
TolFun), or the simplex vertices themselves are within a specified distance (TolX) of each other [16].
Nelder-Mead Algorithm Coefficients
The following table summarizes the standard coefficients used in the Nelder-Mead operations [13].
| Operation | Symbol | Standard Value | Description |
|---|---|---|---|
| Reflection | α (alpha) |
1.0 | Moves the worst point through the centroid of the opposite face. |
| Expansion | γ (gamma) |
2.0 | Pushes further in a promising direction beyond the reflection point. |
| Contraction | Ï (rho) |
0.5 | Pulls the worst point closer to the centroid, either inside or outside. |
| Shrink | σ (sigma) |
0.5 | Reduces the size of the entire simplex towards the best point. |
Nelder-Mead Algorithm Workflow
The following diagram illustrates the logical flow of decisions and operations in a single iteration of the Nelder-Mead method.
The Scientist's Toolkit: Research Reagent Solutions
The following table details key conceptual "reagents" or components essential for conducting an optimization experiment using the Nelder-Mead method.
| Item | Function / Role in the Experiment |
|---|---|
| Objective Function | The function f(x) to be minimized. It is the central metric defining the quality of a candidate solution. |
| Initial Simplex | The starting set of n+1 points in n-dimensional space. Its quality can significantly impact convergence [13] [16]. |
| Reflection Coefficient (α) | Controls the distance the worst point is reflected through the centroid. A value of 1.0 maintains the simplex volume [13]. |
| Expansion Coefficient (γ) | Allows the algorithm to take larger, accelerating steps down a promising valley [13] [15]. |
| Contraction Coefficient (Ï) | Enables the simplex to contract and narrow in on a potential minimum [13]. |
| Termination Tolerance | A predefined threshold for the standard deviation of function values or simplex size that signals the search is complete [16]. |
| Phaseic acid-d4 | Phaseic acid-d4 Stable Isotope|ABA Metabolite |
| L-Biotin-NH-5MP | L-Biotin-NH-5MP, MF:C15H20N4O3S, MW:336.4 g/mol |
Troubleshooting Guide: Common Sequential Simplex Method Issues
Q1: Why is my optimization process stagnating or cycling between the same solutions? This occurs when the simplex collapses or cannot find an improved vertex. To resolve:
- Recalculate all vertices: Ensure objective function values are current.
- Implement a restart: Shrink the simplex towards the current best vertex and restart the search [11].
- Check for tolerance criteria: The process may have converged; define a tolerance for the minimum simplex size or objective function change to terminate correctly [11].
Q2: How do I handle an experiment where one or more factors have natural lower bounds of zero? The sequential simplex method inherently requires all factors (variables) to be non-negative [17].
- No action needed for non-negativity: If your factors are naturally positive (e.g., concentration, temperature in Kelvin), proceed directly.
- Handling bounded variables: For factors with other lower bounds, apply a simple transformation. For a factor ( x ) with a lower bound ( L ), define a new variable ( x' = x - L ). Now, ( x' \geq 0 ) and the simplex method can proceed [17].
Q3: What should I do if the suggested new experiment is practically infeasible or dangerous? The simplex method is a mathematical guide and suggestions must be tempered with practical knowledge.
- Constrain the move: Do not perform the experiment as calculated. Instead, move the new vertex to the nearest feasible point on the boundary of your experimental region.
- Reflect from the second-worst point: Modify the algorithm to reflect away from the second-worst vertex instead of the worst one to propose an alternative point [11].
Frequently Asked Questions (FAQs)
Q: How does the sequential simplex method handle multiple factors without complex statistics?
It uses a geometric approach. For n factors, a simplex with n+1 vertices is constructed in the experimental space. The method then proceeds by moving away from the point with the worst performance through a series of reflections, expansions, and contractions, navigating the factor space based solely on the observed responses without requiring assumptions about the underlying functional form or interactions [11].
Q: What is the main advantage of this method over traditional Design of Experiments (DOE)? Traditional DOE often requires a predefined model (e.g., linear, quadratic) to fit the data and estimate interaction effects. The sequential simplex method is model-free; it does not assume a specific relationship between factors. It efficiently guides the experimenter towards an optimum by reacting to the observed data at the vertices of the simplex, making it highly effective for rapid empirical optimization [11].
Q: When should I not use the sequential simplex method?
- When the objective is to build a predictive model for the entire response surface.
- When the system has a high degree of experimental noise, as this can mislead the simplex moves.
- When you need to understand the precise contribution and interaction of each factor (system understanding vs. empirical optimization) [11].
Q: How do I set up the initial simplex? The initial simplex should be a regular geometric figure. For two factors, it is an equilateral triangle; for three, a tetrahedron. The size of the initial simplex determines the initial step size. A larger simplex coarsely finds the optimum region, while a smaller one performs a finer local search [11].
Experimental Protocol: Implementing the Sequential Simplex Method
The following workflow details the core operational steps of the sequential simplex method for optimizing a process with n factors.
Core Simplex Operations and Rules
The table below defines the key operations used to manipulate the simplex and the rules for accepting new points.
| Operation | Mathematical Calculation | Acceptance Rule |
|---|---|---|
| Reflection | ( R = C + \alpha(C - W) ), where ( C ) is the centroid (excluding W) and ( \alpha > 0 ) (typically 1) [11]. | Accept if ( R ) is better than ( W ) but not better than ( B ). |
| Expansion | ( E = C + \gamma(R - C) ), where ( \gamma > 1 ) (typically 2) [11]. | If ( R ) is better than ( B ), expand to ( E ). Accept ( E ) if it is better than ( R ); otherwise, accept ( R ). |
| Contraction | ( T = C + \beta(W - C) ) or ( T = C + \beta(R - C) ), where ( 0 < \beta < 1 ) (typically 0.5) [11]. | If ( R ) is worse than ( W ), contract to ( T ). If ( T ) is better than ( W ), accept it. Otherwise, proceed to shrinkage. |
| Shrinkage | All vertices except ( B ) are moved: ( Vi^{new} = B + \delta(Vi - B) ), where ( 0 < \delta < 1 ) (typically 0.5) [11]. | Perform if contraction fails. Effectively restarts the search with a smaller simplex around the current best point. |
Key Simplex Parameters
The following table summarizes typical values for the coefficients used in the operations above.
| Parameter | Symbol | Typical Value | Purpose |
|---|---|---|---|
| Reflection | ( \alpha ) | 1.0 | Moves away from the worst-performing region. |
| Expansion | ( \gamma ) | 2.0 | Accelerates progress in a promising direction. |
| Contraction | ( \beta ) | 0.5 | Shrinks the simplex when a reflection fails. |
| Shrinkage | ( \delta ) | 0.5 | Resets the search around the best point to escape a non-productive region. |
The Scientist's Toolkit: Essential Research Reagent Solutions
The sequential simplex method is a mathematical procedure and does not require specific chemical reagents. Its "toolkit" consists of the computational parameters and experimental components required for execution.
| Item / Concept | Function in the Sequential Simplex Method |
|---|---|
| Initial Vertex Matrix | The set of n+1 initial experimental points that define the starting simplex in the n-dimensional factor space [11]. |
| Objective Function | The measurable response (e.g., yield, purity, activity) that the method is designed to maximize or minimize [11]. |
| Reflection Coefficient ((\alpha)) | The multiplicative factor that determines how far the simplex reflects away from the worst vertex. A value of 1.0 is standard [11]. |
| Expansion Coefficient ((\gamma)) | The multiplicative factor that determines how far the simplex expands beyond a successful reflection point to accelerate progress [11]. |
| Contraction Coefficient ((\beta)) | The multiplicative factor that determines how much the simplex contracts towards the centroid when a reflection fails to improve the result [11]. |
| Convergence Tolerance | A pre-defined threshold (for simplex size or objective function change) that signals the optimization is complete [11]. |
| Estriol-d3 | Estriol-d3, MF:C18H24O3, MW:291.4 g/mol |
| Creticoside C | Creticoside C, MF:C26H44O8, MW:484.6 g/mol |
Logical Decision Pathway for a New Experimental Point
The following diagram illustrates the precise logic used to decide whether to accept a reflected point or to trigger an expansion or contraction.
Implementing Sequential Simplex: Step-by-Step Protocols and Real-World Applications
Frequently Asked Questions (FAQs)
What is an Initial Simplex Design and when should I use it?
An initial simplex design is a structured set of experimental runs that serves as the starting point for optimization algorithms, particularly the sequential simplex method. You should use it when you need to efficiently locate optimal conditions ("sweet spots") in multi-factor experiments, especially during scouting studies in fields like bioprocess development or drug formulation. This approach is particularly valuable when you want to reduce experimental costs while still obtaining well-defined operating boundaries compared to traditional Design of Experiments (DoE) methods [18].
How do I determine the starting point and size for a k-factor experiment?
For a k-factor experiment (with k components or factors), the initial simplex is a geometric figure defined by k+1 points. In mixture experiments, these factors are components whose proportions sum to a constant, usually 1 [19] [20].
- Starting Point Selection: The starting point should be within your experimental region of interest. If you have preliminary knowledge suggesting a promising area, start there. Otherwise, a centroid or evenly distributed point is a reasonable default.
- Size Determination: The size determines how much of the experimental space you initially explore. A larger simplex is more exploratory, while a smaller one provides more refined local search. Your choice should balance between broad exploration and resource constraints.
The total number of design points in a {q, m} simplex-lattice design is given by the formula: (q + m - 1)! / (m!(q-1)!) where 'q' is the number of components and 'm' is the number of equally spaced levels for each component [20].
Table: Initial Simplex Design Parameters for Different Factor Counts
| Number of Factors (k) | Geometric Form | Minimum Number of Initial Runs | Design Notation Example |
|---|---|---|---|
| 2 | Triangle | 3 | {3,2} design with 6 points [20] |
| 3 | Tetrahedron | 4 | {3,3} design with 10 points [20] |
| 4 | 5-cell | 5 | {4,2} design |
What is the step-by-step protocol for creating a simplex lattice design?
Protocol: Creating a Three-Factor Simplex Lattice Design [19]
- Define Factors and Constraints: Identify all mixture components. The total proportion of all components must sum to 1 (or 100%).
- Set Factor Levels: Determine the number of equally spaced levels (m) for each component. The proportions for each component will be xi = 0, 1/m, 2/m, ..., 1.
- Generate Design Points: Create all possible combinations of the factors where their proportions follow the levels from step 2 and sum to 1.
- Build Design Table: Assemble the coordinate settings for each experimental run.
- Randomize Run Order: Minimize the effect of unknown nuisance factors by randomizing the order in which you will execute the experimental runs [21].
Example: A {3,2} Simplex Lattice Design [20] This design for three components (q=3) with two levels (m=2) generates the following 6 design points, though the number of experimental observations can be increased with replication.
Table: Design Matrix for a {3,2} Simplex-Lattice
| X1 (Component 1) | X2 (Component 2) | X3 (Component 3) |
|---|---|---|
| 1 | 0 | 0 |
| 0 | 1 | 0 |
| 0 | 0 | 1 |
| 0.5 | 0.5 | 0 |
| 0.5 | 0 | 0.5 |
| 0 | 0.5 | 0.5 |
How do I incorporate process variables into a mixture design?
Many real-world experiments involve both mixture components (which sum to a constant) and process factors (which are independent). You can study them together in a combined design [21].
Protocol: Creating a Design with Process Variables [21]
- Create the Mixture Design: First, generate the simplex lattice design for your mixture components (e.g., a {3,2} simplex lattice).
- Add Process Factors: Specify your process variables (e.g., Temperature, Rate of Extrusion) and their levels (e.g., Low: 10, High: 20).
- Cross the Designs: Conduct the entire mixture design (Step 1) under every possible combination of the process factor levels. For example, with 6 mixture blends and 2 process factors at 2 levels each (4 combinations), your total number of experimental runs would be 6 x 4 = 24.
- Analyze with a Combined Model: Fit a model that includes terms for the mixture components, the process factors, and their interactions.
What are the essential reagent solutions for a simplex-related experiment?
Table: Essential Research Reagent Solutions for Simplex Optimization Experiments
| Reagent / Material | Function in Experiment | Example from Analytical Chemistry [22] |
|---|---|---|
| Standard Stock Solutions | Primary components for creating mixture blends; used as standards for calibration. | 1000 mg L−1 solutions of heavy metals (e.g., Zn(II), Cd(II)) for electrode optimization. |
| Supporting Electrolyte | Provides ionic strength and controls the chemical environment for electrochemical measurements. | 0.1 M acetate buffer solution (pH 4.5). |
| Film Forming Ions | Used to modify and optimize the working electrode surface in electroanalysis. | Bi(III), Sn(II), and Sb(III) ions for forming in-situ film electrodes. |
| Polishing Material | For preparing and maintaining a consistent, clean surface on solid working electrodes. | 0.05 μm Al₂O₃ suspension for polishing glassy carbon electrodes. |
| Real Sample Matrix | Used for method validation and demonstrating applicability to real-world problems. | Tap water prepared in 0.1 M acetate buffer for testing optimized methods. |
Troubleshooting Guides
Problem: The sequential simplex algorithm is not converging to an optimum.
Possible Causes and Solutions:
- Cause 1: Poorly chosen initial simplex size.
- Solution: If the simplex is too large, it may miss fine-grained optimums. If it's too small, progress may be slow. Restart the sequence with a differently sized initial simplex that is appropriate for your expected optimum region [11].
- Cause 2: The initial simplex is located on a flat region of the response surface.
- Solution: If possible, use preliminary knowledge to start in a more responsive region. Alternatively, incorporate curvature into your model if you are using a simplex lattice design for response surface modeling [18].
- Cause 3: The simplex is collapsing or degenerating, losing its volume in the factor space.
- Solution: This is a known issue with the basic sequential simplex method. Implement the modified algorithm by Nelder and Mead, which includes reflection, expansion, contraction, and shrinkage steps to maintain the simplex's geometric properties and prevent collapse [11].
Problem: My analysis shows insignificant model terms or poor model fit.
Possible Causes and Solutions:
- Cause 1: The model is overparameterized for the data collected.
- Solution: Use a model selection procedure. Start by including all linear effects and 2-way interactions, then remove terms with high p-values (e.g., above 0.1) or coefficients with an absolute value less than or equal to 1. Recalculate the model after removing insignificant terms [21].
- Cause 2: The design does not include enough points to estimate the desired model.
- Solution: Ensure your initial design has a sufficient number of runs. A {q, m} simplex-lattice has (q + m - 1)! / (m!(q-1)!) points. For a quadratic model, you need at least enough points to estimate the linear and binary interaction terms [20].
- Cause 3: Important process variables were not considered.
- Solution: If your system is influenced by independent factors like temperature or time, use a combined design that crosses your mixture design with a factorial design for the process variables [21].
Workflow and Signaling Diagrams
Simplex Experiment Initiation Workflow
Mixture Model Interpretation Logic
Troubleshooting Guide: Sequential Simplex Method
This guide addresses common issues you might encounter while using the sequential simplex method to optimize multiple experimental factors.
Q1: The simplex is not moving toward an optimum and seems to be "stagnating" in a suboptimal region. What should I do?
A: This behavior often indicates that the simplex has become stuck on a ridge or is navigating a poorly conditioned response surface.
- Check for Simplex Degeneracy: Ensure that the simplex has not collapsed into a lower-dimensional space. Re-initialize the simplex with a new set of starting points if necessary.
- Verify Movement Logic: Confirm that the reflection, expansion, and contraction operations are correctly implemented. The reflection coefficient (α) is typically 1.0, expansion (γ) is 2.0, and contraction (Ï) is 0.5 [23].
- Consider a Restart: If the simplex has undergone many contractions without significant improvement, halt the current run and restart the procedure from the current best vertex.
Q2: The simplex oscillates between two points instead of converging. How can I resolve this?
A: Oscillation typically occurs when the simplex repeatedly reflects over the same centroid.
- Implement a "Move Rule": Introduce logic to prevent the simplex from returning to a vertex it has just visited. This can break the cycle of reflection between the same points.
- Review Contraction Logic: Ensure that the contraction step is being triggered correctly when a reflected point yields the worst response. A proper contraction should move the simplex away from the oscillatory path [23].
Q3: After a contraction step, the new vertex does not show improvement. What is the next step?
A: Standard contraction might be insufficient if the response surface is complex.
- Perform a "Shrink" or "Total Contraction": If even the contracted vertex does not yield an improvement, a more radical reduction of the simplex size may be necessary. Shrinking involves moving all vertices toward the best-performing vertex, effectively resetting the search on a finer scale around the most promising region [1] [23].
Q4: How do I handle constraints on experimental factors (e.g., pH cannot exceed 14)?
A: The basic simplex method requires modification to handle constrained factors.
- Incorporate Constraint Checks: Before evaluating the response at a new vertex, check if it violates any experimental constraints.
- Apply a Penalty Function: If a proposed vertex violates a constraint, assign it a deliberately poor (e.g., very high for minimization) response value. This will force the simplex to reject this point and trigger a contraction, steering the search back into the feasible region [1].
Q5: The experimental noise is high, leading to unreliable response measurements. How can I make the method more robust?
A: High noise can cause the simplex to move in the wrong direction.
- Replicate Measurements: Instead of a single measurement per vertex, run multiple replicates and use the average response to guide the simplex movements. This reduces the impact of random noise.
- Adjust the Step Size: Use a larger initial simplex size relative to the noise level. This makes the signal from factor changes more detectable compared to the background noise.
Experimental Protocol: Sequential Simplex Optimization
This protocol provides a detailed methodology for conducting an optimization experiment using the sequential simplex method.
1. Objective Definition Define the response variable to be optimized (e.g., yield, purity, activity) and specify whether the goal is to maximize or minimize it. Clearly identify all independent factors (e.g., temperature, concentration, pressure) that will be adjusted.
2. Initial Simplex Construction The initial simplex is a geometric figure with k+1 vertices, where k is the number of factors being optimized.
- Starting Point: Select a feasible starting point (vertex) based on prior knowledge, designated as Vâ‚.
- Factor Scaling: Ensure all factors are on a comparable scale (e.g., by normalizing) to prevent one factor from dominating the movement.
- Generate Remaining Vertices: The other k vertices are generated based on a specified step size for each factor. A common approach is to use the variable-size matrix to construct the initial simplex [23].
3. The Optimization Cycle The following workflow is repeated until a termination criterion is met (e.g., the simplex size becomes smaller than a pre-defined threshold, or the response improvement plateaus).
4. Decision Rules and Movement Logic The response at the reflected vertex (R_reflect) determines the next action, according to the logic in the table below.
| New Vertex | Condition Check | Outcome & Action |
|---|---|---|
| Reflect | Always the first step after ranking and finding the centroid. | Base case for decision-making [23]. |
| Expand | If Rreflect is better than the current best response (Vbest). | The direction is highly favorable. Calculate Vexpand = Vcentroid + γ*(Vreflect - Vcentroid). Evaluate Rexpand. If Rexpand > Rreflect, replace Vworst with Vexpard. Otherwise, use Vreflect [23]. |
| Contract | If Rreflect is worse than or equal to the response at Vsecondworst (or another vertex besides Vworst). | The reflection went too far. Calculate Vcontract = Vcentroid + Ï*(Vworst - Vcentroid). Evaluate Rcontract. If Rcontract > Rworst, replace Vworst with V_contract. Otherwise, proceed to a shrink step [23]. |
| Shrink | If the point from contraction (Vcontract) is not better than Vworst. | The entire simplex must be reduced. Move all vertices (Vi) toward the best vertex (Vbest) according to the rule: Vinew = Vbest + σ*(Viold - Vbest), where σ is the shrink coefficient (typically 0.5) [1] [23]. |
The Scientist's Toolkit: Research Reagent Solutions
| Item Name | Function / Role in Optimization |
|---|---|
| High-Throughput Screening (HTS) Assay Kits | Provides a reliable, standardized method for rapidly measuring the biological response (e.g., enzyme activity, cell viability) at each experimental vertex. |
| Statistical Software (e.g., R, Python with SciPy) | Used to implement the simplex algorithm's logic, perform calculations (e.g., centroid), and visualize the path of the simplex across the response surface [24]. |
| Design of Experiments (DoE) Software | Platforms like Ax or other custom solutions can help manage the sequential experiments, track responses, and sometimes directly implement optimization algorithms [24]. |
| Central Composite Design (CCD) | Though a classical method, a CCD can be used in later stages to build a precise local model of the response surface around the optimum found by the simplex, confirming the result [23]. |
| Parameter Tuning Platform (e.g., Ax) | An adaptive experimentation platform that can be used to validate simplex results or handle optimization problems with a very high number of factors where Bayesian optimization might be more efficient [24]. |
| Hypericin (Standard) | Hypericin (Standard), MF:C30H16O8, MW:504.4 g/mol |
| Sibiricine | Sibiricine, CAS:24181-66-6, MF:C20H17NO6, MW:367.4 g/mol |
Frequently Asked Questions (FAQs)
Q: How is the sequential simplex method different from other optimization approaches like Bayesian optimization?
A: The sequential simplex is a direct search method that uses simple geometric rules (reflect, expand, contract) and does not require building a statistical model of the entire response surface. It is conceptually straightforward and efficient with a small number of factors. In contrast, Bayesian optimization is a model-based method that uses a probabilistic surrogate model (like a Gaussian Process) and an acquisition function to guide experiments. It is often more efficient in high-dimensional spaces or when experiments are extremely expensive [24] [23].
Q: What are the typical values for the reflection (α), expansion (γ), and contraction (Ï) coefficients?
A: The most standard set of coefficients is α = 1.0, γ = 2.0, and Ï = 0.5. These values have been found to provide a robust balance between aggressive movement toward an optimum (expansion) and cautious refinement (contraction) [23].
Q: When should I terminate a simplex optimization run?
A: Termination is usually based on one or more of the following criteria:
- The difference in response values between the best and worst vertices falls below a pre-set tolerance.
- The simplex has become very small (the distance between vertices is below a threshold).
- A predetermined maximum number of experiments has been conducted.
Q: Can the simplex method handle more than three factors effectively?
A: Yes, the simplex method can be applied to k factors, forming a k+1 vertex polytope in k-dimensional space. However, as the number of factors increases, the number of experiments required can grow, and the method may become less efficient compared to some model-based techniques. It is generally most practical for optimizing up to about five or six factors [1] [23].
Troubleshooting Guides
Low Protein Yield
- Problem: Insufficient production of the target recombinant protein.
- Possible Causes & Solutions:
| Cause | Diagnostic Method | Solution |
|---|---|---|
| Toxic protein to host cells | SDS-PAGE analysis post-induction; monitor cell growth arrest. [25] | Use specialized E. coli strains like C41(DE3) or C43(DE3) designed for toxic proteins. [25] |
| Inefficient transcription/translation | Check plasmid sequence for errors and codon usage. [25] | Use codon-plus strains like Rosetta; optimize induction conditions (IPTG concentration, temperature, timing). [25] |
| Protein degradation | Western blot showing smeared or disappearing bands. [25] | Use protease-deficient host strains (e.g., BL21); lower induction temperature; add protease inhibitors during lysis. [25] |
| Insufficient culture aeration/nutrient depletion | Monitor OD600 and growth curve. [25] | Increase shaking speed; reduce culture volume; scale up culture volume; use rich media like Terrific Broth. [25] |
Poor or Inefficient Biotinylation
- Problem: The target protein is expressed but is not biotinylated, or biotinylation efficiency is low.
- Possible Causes & Solutions:
| Cause | Diagnostic Method | Solution |
|---|---|---|
| BirA biotin ligase activity or concentration is limiting | Use streptavidin-AP Western blot to detect biotinylation. [26] | Co-express BirA ligase with your target protein; ensure adequate biotin and ATP in the culture medium. [27] |
| Inaccessible biotin acceptor peptide (AviTag) | Confirm protein sequence and AviTag placement. | Ensure the AviTag is located in a flexible, solvent-accessible region of your protein, often at the N- or C-terminus. |
| Sub-optimal in vivo reaction conditions | Measure biotinylation efficiency in vitro. [26] | Supplement culture media with excess biotin (e.g., 50 μM); induce BirA expression before target protein induction; use a mutant BirA (R118G) with promiscuous activity (use with caution). [26] |
Low Protein Solubility
- Problem: The target protein is produced but forms inclusion bodies (insoluble aggregates).
- Possible Causes & Solutions:
| Cause | Diagnostic Method | Solution |
|---|---|---|
| Aggregation during high-level expression | Solubility analysis via centrifugation and SDS-PAGE of soluble vs. insoluble fractions. [25] | Lower induction temperature (e.g., 16-25°C); reduce inducer (IPTG) concentration; use fusion tags like MBP or SUMO to enhance solubility. [25] |
| Lack of proper folding partners | Compare solubility in different E. coli strains. [25] | Use strains engineered for disulfide bond formation (e.g., Shuffle T7) or co-express molecular chaperones. [25] |
| Incorrect or harsh lysis conditions | Visualize protein localization. [25] | Use gentle lysis methods; screen different lysis buffers with varying salt, pH, and detergent concentrations. [25] |
Frequently Asked Questions (FAQs)
Q1: What is the most suitable E. coli strain for producing a biotinylated protein that requires disulfide bonds for activity? The Shuffle strain series is an excellent choice. These strains are engineered to promote the correct formation of disulfide bonds in the cytoplasm by expressing disulfide bond isomerase (DsbC) and are deficient in proteases, enhancing the stability of your recombinant protein. [25]
Q2: My biotinylated protein is expressed in inclusion bodies. Should I attempt to refold it or change my strategy? You can do either, but a strategy change is often more efficient. First, attempt to refold the protein from inclusion bodies using denaturing agents like urea or guanidine hydrochloride, followed by gradual dialysis into a native buffer. Alternatively, optimize expression for solubility by switching to a lower induction temperature (18-25°C), using a solubility-enhancing fusion tag (like MBP or SUMO), or trying a different E. coli host strain. [25]
Q3: How can I confirm that my protein is successfully biotinylated? The most common and reliable method is a Western blot. [26] After separating your protein samples via SDS-PAGE, transfer them to a membrane and probe with streptavidin conjugated to a detection molecule (e.g., horseradish peroxidase or alkaline phosphatase). Streptavidin binds with extremely high affinity to biotin, allowing you to visualize only the biotinylated proteins. [26]
Q4: Can I perform high-throughput screening for optimizing biotinylated protein production? Yes, high-throughput screening (HTS) is feasible and recommended for multi-factor optimization. You can perform cloning, expression, and initial screening in 96-well or 384-well microplates. [28] For the expression step, technologies like the Vesicle Nucleating peptide (VNp) can be used to export functional proteins into the culture medium in a multi-well format, simplifying purification and analysis. [28] This setup is ideal for testing many different conditions in parallel.
Q5: What is the role of the sequential simplex method in this optimization process? The sequential simplex method is a powerful statistical optimization tool for experiments with multiple variables. [29] In the context of optimizing protein production, you can use it to efficiently navigate factors like temperature, inducer concentration, media composition, and biotin concentration. [29] Unlike testing one factor at a time, the simplex method moves towards an optimum based on the results of previous experiments, often requiring fewer total experiments to find the ideal condition combination. [29]
Research Reagent Solutions
The following table lists key reagents and materials essential for the experiments described in this case study.
| Item | Function/Application in the Experiment |
|---|---|
| E. coli Strains (BL21(DE3), Shuffle, Rosetta) | Engineered host organisms for recombinant protein expression, offering benefits like protease deficiency, enhanced disulfide bond formation, or supplementation of rare tRNAs. [25] |
| pET Expression Vectors | Plasmid vectors containing a strong T7 promoter, used to clone the gene of interest and drive high-level protein expression in E. coli. [25] |
| BirA Biotin Ligase | The enzyme that catalyzes the attachment of biotin to a specific lysine residue within the AviTag sequence on the target protein. [26] [27] |
| Biotin | The essential co-factor and substrate for the BirA enzyme. Must be supplemented in the culture medium for in vivo biotinylation. [26] |
| Ni-NTA Affinity Resin | For purifying recombinant proteins that have been fused with a hexahistidine (6xHis) tag. The resin binds the His-tag with high specificity. [25] |
| Strep-Tactin Magnetic Beads | Beads that bind with high affinity to biotin. Used to rapidly and efficiently purify biotinylated proteins, ideal for automated or high-throughput workflows. [27] |
| VNp (Vesicle Nucleating Peptide) Tag | A peptide tag that, when fused to a recombinant protein, promotes its export from E. coli into extracellular vesicles, simplifying purification and enhancing stability. [28] |
Experimental Workflow Diagrams
Workflow for Standard Production and Optimization
High-Throughput Screening Workflow
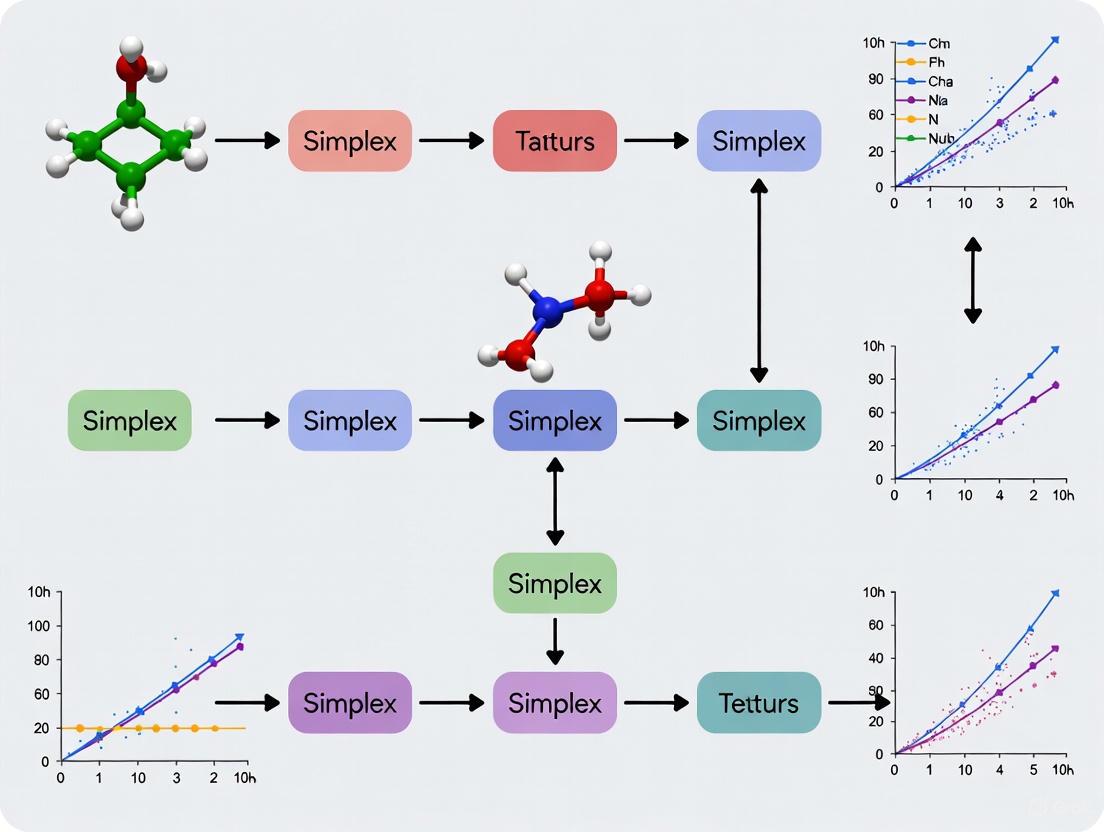
The sequential simplex method is a powerful, practical multivariate optimization technique used in analytical method development to efficiently find the optimal conditions for a process by considering multiple variables simultaneously. Unlike univariate optimization, which changes one factor at a time, the simplex method evaluates all factors concurrently, allowing researchers to identify interactions between variables and reach optimum conditions with fewer experiments [12].
In this case study, we demonstrate how the sequential simplex method was successfully employed to optimize chromatographic and spectroscopic methods for pharmaceutical analysis. This approach is particularly valuable in analytical chemistry for optimizing instrumental parameters, developing robust analytical procedures, and ensuring good analytical characteristics such as higher sensitivity and accuracy [12]. The method works by displacing a geometric figure with k + 1 vertexes (where k equals the number of variables) through an experimental field toward an optimal region, with movements including reflection, expansion, and contraction to navigate the response surface efficiently [12].
Experimental Protocols & Workflows
Sequential Simplex Optimization: A Step-by-Step Protocol
The following workflow illustrates the core decision-making process of the sequential simplex method:
Detailed Experimental Protocol for Simplex Optimization
Materials: Standard analytical reagents, chromatographic reference standards, appropriate instrumentation (HPLC, GC, or spectrophotometer), data collection software.
Procedure:
- Define Variables and Responses: Select critical factors to optimize (e.g., mobile phase composition, pH, temperature, flow rate) and measurable responses (e.g., resolution, peak symmetry, sensitivity) [12].
- Establish Initial Simplex: Create an initial geometric figure with k+1 vertexes, where k is the number of variables being optimized. For two variables, this forms a triangle; for three variables, a tetrahedron [12].
- Run Experiments: Conduct experiments at each vertex of the initial simplex and measure the responses.
- Calculate New Vertex:
- Reflection: Calculate the reflection point (R) of the worst vertex (W) through the centroid of the remaining vertexes using the formula: R = P + α(P - W), where P is the centroid and α is the reflection coefficient (typically 1.0) [12].
- Expansion: If the reflection point yields better results than the current best vertex, calculate an expansion point (E) using: E = P + γ(P - W), where γ is the expansion coefficient (typically >1.0) [12].
- Contraction: If the reflection point is worse than the second-worst vertex, calculate a contraction point (C) using: C = P + β(P - W), where β is the contraction coefficient (typically 0.5) [12].
- Iterate: Replace the worst vertex with the new vertex and repeat the process until the simplex converges on the optimum conditions [12].
- Verify: Confirm optimal conditions with replicate experiments and validate the method according to regulatory guidelines [30].
Case Study: Fast-Dissolving Tablet Formulation Development
In a practical application from pharmaceutical research, the sequential simplex method was employed to develop fast-dissolving tablets of clozapine [31]. Researchers selected microcrystalline cellulose and polyplasdone as the two critical formulation variables and evaluated responses including disintegration time, hardness, and friability. The success of formulations was evaluated using a total response equation generated in accordance with the priority of the response parameters [31]. Based on response rankings, the simplex sequence continued through reflection, expansion, or contraction operations until a desirable disintegration time of less than 10 seconds with adequate hardness was achieved [31].
Chromatographic Method Development Compliance with USP <621>
When developing chromatographic methods for pharmaceutical applications, compliance with regulatory standards is essential. The United States Pharmacopeia (USP) General Chapter <621> Chromatography provides mandatory requirements for chromatographic analysis in regulated Good Manufacturing Practice (GMP) laboratories [30]. Key considerations include:
System Suitability Testing (SST) Requirements:
- System Sensitivity: Signal-to-noise (S/N) ratio measurements are required when determining impurities, with the limit of quantification (LOQ) based on a S/N of 10 [30].
- Peak Symmetry: A general requirement for peak symmetry between 0.8-1.8 has been added to the updated USP <621> chapter [32].
- Resolution Calculation: The definition and calculation of resolution is now based on peak width at half height rather than at the baseline [32].
Allowed Adjustments: The harmonized USP <621> standard allows certain adjustments to chromatographic systems without requiring full revalidation, including changes to injection volume, mobile phase composition, pH, and concentration of salts in buffers, provided system suitability requirements are met [32] [30].
Troubleshooting Guides & FAQs
Chromatography Troubleshooting Guide
Table 1: Common Chromatography Issues and Solutions
| Problem | Possible Causes | Troubleshooting Steps | Prevention Tips |
|---|---|---|---|
| No peaks or only solvent peak [33] | - Column installed incorrectly- Split ratio too high (capillary GC)- Large leak (septum, column connections, detector)- Detector settings incorrect- Sample issues | 1. Verify column installation and connections2. Check split ratio settings3. Perform leak check4. Verify detector configuration5. Test with known standard mixture | - Follow manufacturer's column installation guidelines- Regularly change septa and check fittings- Use retention gaps where appropriate |
| Peak fronting [33] | - Column overload- Inappropriate injector liner- Incorrect injection technique | 1. Reduce injection volume or sample concentration2. Change inlet liner3. Verify injection technique and parameters | - Ensure sample concentration is within linear range- Select proper liner for application- Follow established injection protocols |
| Peak tailing [33] | - Active sites in column or system- Column contamination- Incorrect mobile phase pH- Sample interaction with system components | 1. Condition column properly2. Use peak tailing reagents in mobile phase3. Adjust mobile phase pH4. Replace column if severely degraded | - Use appropriate guard columns- Filter samples and mobile phases- Regular column maintenance and cleaning |
| Poor resolution [32] [30] | - Incorrect mobile phase composition- Column deterioration- Temperature inappropriate- Flow rate too high | 1. Optimize mobile phase composition2. Evaluate column performance with test mix3. Adjust temperature parameters4. Reduce flow rate | - Monitor system suitability parameters regularly- Establish column performance tracking- Follow manufacturer's recommended conditions |
Frequently Asked Questions (FAQs)
Q: When should I use sequential simplex optimization instead of other optimization methods? A: Sequential simplex is particularly valuable when you need to optimize multiple factors simultaneously without requiring complex mathematical-statistical expertise [12]. It's especially useful for optimizing automated analytical systems and when dealing with systems where variable interactions are significant. The method provides a practical approach that can be easily implemented and understood by researchers with varying levels of statistical background.
Q: What are the most critical changes in the updated USP <621> chromatography chapter? A: The most significant changes include: (1) New requirements for system sensitivity (signal-to-noise ratio) with LOQ based on S/N of 10; (2) General peak symmetry requirements between 0.8-1.8; (3) Resolution calculation based on peak width at half height; (4) Expanded allowed adjustments for gradient elution methods; and (5) Replacement of "disregard limit" with "reporting thresholds" [32] [30]. These changes became fully effective May 1, 2025.
Q: Why do I see only solvent peaks after installing a new GC column? A: This common issue can have multiple causes [33]. First, verify proper column installation and check for leaks at injector septa and column connections. Second, confirm your split ratio isn't set too high for capillary columns. Third, check detector settings and ensure they're configured correctly. Finally, test with a known standard mixture to verify system performance. Always condition new columns according to manufacturer specifications before use.
Q: How do I calculate resolution according to the updated USP <621> guidelines? A: The updated USP <621> chapter has modified the resolution calculation to be based on peak width at half height rather than at the baseline [32]. This provides a more consistent measurement, particularly for peaks of different shapes or those with tailing. Ensure your chromatography data system software is updated to use the current calculation method to maintain compliance.
Q: Can I adjust my chromatographic method parameters without revalidation? A: Yes, within specific limits outlined in USP <621> [32] [30]. The harmonized standard allows adjustments including mobile phase composition, pH, concentration of salts in buffers, application volume (TLC), and injection volume, provided system suitability requirements are met. For gradient elution methods, adjustments to particle size and injection volume are now permitted. However, any adjustments must be documented and verified to ensure they don't compromise method validity.
Research Reagent Solutions & Essential Materials
Table 2: Key Research Reagents and Materials for Analytical Method Development
| Reagent/Material | Function/Application | Usage Notes |
|---|---|---|
| Microcrystalline Cellulose [31] | Pharmaceutical excipient used in formulation development | Used as a variable in simplex optimization of fast-dissolving tablets; provides bulk and compression properties |
| Polyplasdone [31] | Superdisintegrant in tablet formulations | Critical factor in optimizing disintegration time in pharmaceutical formulations |
| FAMEs Standard [33] | Fatty Acid Methyl Esters mixture for GC calibration and column testing | Used for testing GC system performance; particularly important after column installation or maintenance |
| Chromatographic Reference Standards [30] | Qualified materials for system suitability testing | Essential for verifying chromatographic system performance and compliance with USP <621> requirements |
| Appropriate Buffer Systems [32] [30] | Mobile phase components for pH control | Critical for maintaining consistent retention times and peak shapes; pH adjustments allowed under USP <621> within limits |
Method Validation & Regulatory Compliance
The following workflow illustrates the relationship between optimization, method development, and regulatory compliance:
Implementing Updated USP <621> Requirements
The updated USP <621> chromatography chapter introduces specific requirements that laboratories must implement for regulatory compliance [32] [30]:
System Sensitivity Requirements:
- Signal-to-noise (S/N) ratio measurements are required when determining impurities at or near limits of quantification
- The Limit of Quantification (LOQ) is based on a S/N of 10 and is related to the monograph specifications
- S/N ratio should be measured using pharmacopoeial reference standards, not samples
- This requirement applies specifically to impurity determination, not main peak analysis
Peak Symmetry Specifications:
- General requirement for peak symmetry between 0.8-1.8
- Must be measured during system suitability testing
- Provides indication of column performance and appropriate mobile phase conditions
Documentation and Adjustment Protocols:
- Laboratories must document all adjustments made to chromatographic conditions
- Allowed adjustments include changes to injection volume, mobile phase composition, pH, and concentration of salts in buffers
- Changes to gradient elution methods are now permitted without full revalidation, provided system suitability requirements are met
The sequential simplex method provides an efficient, practical approach for optimizing multiple experimental factors in analytical method development. By systematically navigating the experimental response surface, researchers can identify optimal conditions for chromatographic and spectroscopic methods while considering variable interactions. When combined with understanding of regulatory requirements such as USP <621>, this optimization strategy supports the development of robust, reliable analytical methods suitable for pharmaceutical applications and other regulated environments. The troubleshooting guides and FAQs presented in this technical support center address common practical challenges encountered during method development and implementation, providing researchers with actionable solutions to maintain analytical system performance and regulatory compliance.
Frequently Asked Questions (FAQs)
1. What are the default values for reflection, expansion, and contraction coefficients in the simplex method, and when should I deviate from them? The widely accepted default coefficients for the sequential simplex method are a reflection coefficient of 1.0, a contraction coefficient of 0.5, and an expansion coefficient between 2.0 and 2.5 [34]. These values are nearly optimal for many test functions. You should consider using a larger expansion coefficient (e.g., 2.2–2.5) when you need the simplex to search a larger area of the response surface, as this can resemble the effect of repetitive expansion and potentially increase the speed of optimization [34]. However, unlimited repetitive expansion can be less successful for complex functions and must be managed with constraints on degeneracy [34].
2. My simplex optimization keeps failing to converge. What are the primary convergence criteria, and how can I adjust them? Convergence is typically checked by comparing the function value between the points of the current simplex and the corresponding points from the previous iteration [35]. The algorithm often uses two main criteria: an absolute difference threshold and a relative difference threshold [35]. Convergence is achieved when the change in the objective function value between iterations falls below one of these thresholds. If your simplex is failing to converge, you can adjust the convergence checker by specifying custom absolute and relative threshold values. Tighter thresholds will require more iterations but may yield a more precise optimum, whereas looser thresholds may stop prematurely [35].
3. What is simplex degeneracy, and how can I prevent it from causing false convergence? Degeneracy occurs when the simplex vertices become computationally dependent, often due to repeated failed contractions, which can prevent the simplex from progressing toward the true optimum [34]. A key sign is the simplex failing to move or converging to a non-optimal point. To prevent this, implement a constraint on the degeneracy of the simplex, typically by monitoring and controlling the angles between its edges [34]. Furthermore, combining this constraint with a translation of the repeated failed contracted simplex has proven to be a more reliable method for finding the optimum region [34].
4. How should I handle experimental points that fall outside my predefined variable boundaries? A robust approach is to correct the vertex located outside the boundary back to the boundary itself [34]. Simply assigning an unfavorable response value to points outside the boundaries, as done in the basic and modified simplex methods, can be inefficient, especially when the optimum is near a boundary. The correction method improves both the speed and the reliability of locating the optimum [34].
5. What is the recommended way to define the initial simplex to ensure a successful optimization? The initial simplex should be carefully configured to adequately probe the experimental domain. One common method is to define it from a starting point and a set of step variations. You provide a starting point (an array of initial factor levels) and a delta vector [35]. The initial simplex is then generated by adding each coordinate of the delta vector to the corresponding coordinate of the starting point, creating a simplex with edges parallel to the factor axes. Ensure that no value in your delta vector is zero, or the simplex will have a lower dimension and the optimization will fail [35].
Troubleshooting Guides
Problem: Slow or Inefficient Convergence
Possible Causes and Solutions:
- Cause 1: Oversized or undersized initial simplex.
- Solution: Adjust the initial step configuration (
deltaParam). A very small simplex will take many steps to move across the factor space, while a very large one may overshoot the optimum. You may need to scale your step sizes relative to the expected scale of each factor.
- Solution: Adjust the initial step configuration (
- Cause 2: Unfavorable response surface geometry.
- Solution: Consider implementing a variable-size simplex approach. These methods contain additional rules for adapting the size (and sometimes shape) of the simplex based on performance. A larger simplex moves fast towards the optimum, while a smaller one can refine the search close to the optimum [36].
- Cause 3: Repetitive expansions causing instability.
- Solution: While a larger expansion coefficient can help, unlimited repetitive expansion can be counterproductive. Combine a constraint on degeneracy with a constraint on the response increasement for each repetitive expansion to stabilize the process [34].
Problem: Convergence to a Non-Optimal Point (False Convergence)
Possible Causes and Solutions:
- Cause 1: Simplex degeneracy.
- Solution: As outlined in the FAQs, implement degeneracy control. Calculate the degeneracy (e.g., based on angles within the simplex) and apply a constraint to prevent the simplex from collapsing. Performing these calculations only when a vertex has been successfully replaced can further improve efficiency [34].
- Cause 2: Poor boundary handling.
- Solution: Replace the strategy of assigning bad values to out-of-bounds vertices. Instead, use a boundary correction method that moves any vertex outside a boundary back to the boundary line/surface. This is particularly critical when the true optimum is located on or very near a constraint boundary [34].
- Cause 3: The algorithm is stuck in a local optimum.
- Solution: The simplex method is generally designed to find a local optimum. If your response surface is suspected to have multiple optima, use a "classical" approach or a global search technique first to identify the general region of the global optimum. The simplex method can then be used for fine-tuning in that region [37].
Problem: Algorithm Termination Due to Excessive Iterations or Evaluations
Possible Causes and Solutions:
- Cause 1: Overly strict convergence criteria.
- Solution: Loosen the absolute and relative thresholds in your
SimpleScalarValueCheckerobject. The default values are extremely small; increasing them will allow the algorithm to stop sooner [35].
- Solution: Loosen the absolute and relative thresholds in your
- Cause 2: Maximum iteration or evaluation limit is too low.
- Solution: Increase the maximum limits using the
setMaxIterations()andsetMaxEvaluations()methods. This is essential for complex problems with many factors or noisy response functions that require more steps to stabilize [35].
- Solution: Increase the maximum limits using the
- Cause 3: The objective function is computationally expensive.
- Solution: If each function evaluation is slow, start with a larger initial simplex and looser convergence criteria to find a reasonably good solution faster. You can then restart the optimization from that point with a smaller simplex and tighter criteria for final refinement.
Experimental Protocols & Data Presentation
Table 1: Standard and Recommended Simplex Coefficients
This table summarizes the core parameters that control the movement of the simplex [34] [35].
| Coefficient | Operation | Default Value | Recommended Range | Function |
|---|---|---|---|---|
| Reflection (Ï) | Reflection | 1.0 | 1.0 | Generates a new vertex by reflecting the worst point through the centroid of the remaining points. |
| Expansion (χ) | Expansion | 2.0 | 2.2 - 2.5 | Extends the reflection further if the reflected point is much better, allowing the simplex to move faster. |
| Contraction (γ) | Contraction | 0.5 | 0.5 | Shrinks the simplex towards a better point when reflection fails, helping to refine the search. |
| Shrinkage (σ) | Shrinkage | 0.5 | 0.5 | A global contraction that shrinks the entire simplex towards the best vertex when all else fails. |
Table 2: Configuration of Convergence Checker
This table outlines the parameters for the SimpleScalarValueChecker, which determines when the optimization stops [35].
| Parameter | Description | Default Value | Adjustment Guidance |
|---|---|---|---|
| Absolute Threshold | The absolute difference in the function value between two iterations. | Very small (system-dependent) | Increase for earlier termination on flat regions; decrease for higher precision. |
| Relative Threshold | The relative difference (relative to the current function value) between two iterations. | Very small (system-dependent) | Useful for scaling the convergence check when the function value is very large or small. |
| Max Iterations | The maximum number of algorithm iterations allowed. | Largest system integer | Set to a practical number (e.g., 1000) to prevent infinite loops. |
| Max Evaluations | The maximum number of function evaluations allowed. | Largest system integer | Critical to set if each experiment is costly or time-consuming. |
Protocol: Implementing a Robust Simplex Optimization
This methodology is framed within research for optimizing multiple experimental factors, such as chemical processes or analytical methods [34] [37].
Pre-Optimization Setup:
- Define the Objective Function: Clearly specify the response to be maximized (e.g., yield, sensitivity) or minimized (e.g., impurities, cost).
- Select Factors and Ranges: Identify all continuously variable factors to be optimized and their feasible boundaries.
Algorithm Initialization:
- Set Coefficients: Initialize the Nelder-Mead optimizer with the standard coefficients (Ï=1.0, γ=0.5, χ=2.0-2.5, σ=0.5) [34] [35].
- Define Initial Simplex: Choose a sensible starting point based on prior knowledge. Use the
setStartConfigurationmethod with a non-zero delta vector to generate the initial simplex [35]. - Configure Convergence: Create a
SimpleScalarValueCheckerwith appropriate absolute and relative thresholds for your specific problem. SetMaxIterationsandMaxEvaluationsto safe limits [35].
Execution and Monitoring:
- Run the optimizer and record the path of the simplex.
- Monitor the number of function evaluations and iterations.
Post-Optimization Analysis:
- Validate the Result: Confirm the optimal conditions by running a confirmation experiment.
- Model the Region (Optional): Once the optimum is found, use a response surface design (e.g., Central Composite Design) in the region of the optimum to build a model and understand factor interactions [37] [36].
Workflow Visualization
Simplex Optimization Logic
The Scientist's Toolkit: Research Reagent Solutions
Table 3: Essential Computational Tools for Simplex Optimization
| Item | Function in Optimization |
|---|---|
| NelderMead Optimizer Class | The core algorithm engine that executes the sequential simplex logic, including reflection, expansion, and contraction operations [35]. |
| MultivariateRealFunction Interface | A required template for defining your custom objective function, which calculates the system's response (e.g., yield, purity) for any given set of factor levels [35]. |
| GoalType Object (MINIMIZE/MAXIMIZE) | A simple switch that configures the optimizer to either minimize or maximize the objective function [35]. |
| RealPointValuePair Object | A data structure used to store the results of the optimization, containing both the coordinates of the optimal point and the optimal function value [35]. |
| SimpleScalarValueChecker | The convergence watchdog that monitors changes in the objective function between iterations and decides when the optimum has been sufficiently approached [35]. |
| Test Functions (e.g., Himmelblau) | Well-understood mathematical functions with known optima, used to validate and benchmark the performance of the optimization algorithm before applying it to real experimental data [35]. |
| Wangzaozin A | Wangzaozin A |
Navigating Practical Challenges: Noise Management, Convergence Issues, and Performance Optimization
Frequently Asked Questions (FAQs)
Q1: What are the primary sources of noise in biological experiments, and how do they differ? Biological noise originates from two main sources, classified as intrinsic and extrinsic noise. Intrinsic noise refers to the stochastic fluctuations inherent to biochemical reactions within a single cell, such as random transcription factor binding dynamics or the stochastic timing of transcription and translation, which lead to variability in mRNA and protein levels even in genetically identical cells under identical conditions [38] [39]. Extrinsic noise arises from cell-to-cell variations in global cellular factors, such as differences in cell cycle stage, metabolic state, or levels of transcriptional/translational machinery, which cause correlated fluctuations in the expression of multiple genes [40] [41]. The key difference lies in the scope: intrinsic noise is gene-specific and unpredictable at the single-cell level, while extrinsic noise introduces population-level heterogeneity by affecting many cellular components simultaneously [39].
Q2: How can I determine if my experiment has sufficient replication to overcome biological noise? Adequate replication is determined more by the number of biological replicates than by the depth of technical measurements (e.g., sequencing depth) [42]. To optimize sample size, researchers should perform a power analysis, which calculates the number of biological replicates needed to detect a specific effect size with a given probability. This analysis requires defining the effect size (the minimum biologically relevant change), the within-group variance (often estimated from pilot data or published studies), the false discovery rate, and the desired statistical power [42]. Furthermore, it is critical to avoid pseudoreplication, which occurs when measurements are not statistically independent (e.g., treating multiple technical measurements from the same biological sample as independent replicates), as this artificially inflates sample size and leads to false positives [42].
Q3: Can biological noise ever be beneficial for an experiment or a biological system? Yes, noise is not always a detriment and can be functionally important. According to the Constrained Disorder Principle (CDP), an optimal range of noise is essential for the proper functioning and adaptability of all biological systems [43]. For example, stochastic phenotypic variation in an isogenic population can serve as a bet-hedging strategy, allowing a subset of cells to survive sudden environmental changes [41]. In development, noise can drive cell fate decisions, and in immune responses, stochastic production of cytokines can enhance the plasticity of T cells [43] [39]. The goal in experimentation and system design is often to manage and constrain noise within functional boundaries, not to eliminate it entirely [43].
Q4: What is the relationship between promoter architecture and transcriptional noise? Specific DNA sequence features in gene promoters are strongly associated with the level of transcriptional noise. Genes with TATA-box-containing promoters typically show high variability in transcript abundance across a population of cells [38] [39]. This architecture may be selectively advantageous for genes that need to respond rapidly to environmental stresses [38]. Conversely, the presence of CpG islands (CGIs) in promoter regions is associated with reduced transcriptional variability, promoting stable gene expression [38]. The number of transcription factor binding sites (TFBSs) and transcriptional start sites (TSSs) also modulates noise, with more TFBSs increasing and more TSSs decreasing variability [38].
Troubleshooting Guides
Issue 1: High Cell-to-Cell Variability in Gene Expression Measurements
Problem: Single-cell data (e.g., from scRNA-seq or live-cell imaging) shows high variability in gene expression, obscuring the signal of interest. Solution:
- Characterize Noise Type: Determine if variability is primarily intrinsic or extrinsic. This can be done experimentally using dual-reporter systems, where two identical promoters in the same cell drive different fluorescent proteins. Correlated expression indicates extrinsic noise, while uncorrelated expression points to intrinsic noise [38] [41].
- Modulate Expression Strategy: To reduce extrinsic noise, use genomically integrated constructs with identical, strong promoters, which couple the expression of circuit components and minimize population-level variation [40]. To mitigate intrinsic noise, consider increasing gene dosage or using promoters with features that lower noise, such as those lacking TATA boxes [38].
- Employ Computational Tools: Use specialized bioinformatics tools for single-cell data analysis, such as those that identify Differentially Distributed Genes (DDGs) or models like MMIDAS, which are designed to distinguish true biological variation from technical noise [43].
Issue 2: Unstable or Inconsistent Output from a Synthetic Biological Circuit
Problem: A genetically encoded oscillator (GEO) or other synthetic circuit exhibits a wide distribution of dynamic behaviors (e.g., frequency, amplitude) within a clonal population. Solution:
- Map the Phase Portrait: Systematically vary the expression levels of key circuit components (e.g., ATPase and Activator for a GEO) and measure the resulting output (e.g., oscillation frequency and amplitude). This creates a phase portrait that defines the operational boundaries of the circuit [40].
- Exploit Noise Principles: Based on the phase portrait, design an expression strategy that uses noise to your advantage. For instance, if frequency is controlled by a ratio of components (and is robust to intrinsic noise) while amplitude is set by absolute abundance (and is sensitive to extrinsic noise), you can design a system where these properties are tuned independently [40].
- Implement Noise-Filtering Expression Strategies: Use expression architectures that filter out undesirable noise. For example, a genomically integrated, single-copy circuit can drastically reduce extrinsic noise and produce highly consistent oscillatory behavior compared to a multi-copy plasmid-based system [40].
Issue 3: Low Signal-to-Noise Ratio (SNR) in Strain or Biochemical Measurements
Problem: The measured signal from a strain gauge or similar biochemical assay is weak and obscured by electrical or environmental noise. Solution:
- Increase Signal Amplitude: For strain measurements, carefully increase the excitation voltage of the Wheatstone bridge. This directly improves the SNR, but must be balanced against self-heating effects that can cause instability [44].
- Shield and Shorten Cables: Reduce the length of lead wires and shield them from external electromagnetic fields by enclosing them in a grounded chassis to minimize 50/60 Hz power line interference [44].
- Use Appropriate Hardware: Select a measurement device with a large dynamic range and high Effective Number of Bits (ENOB) to minimize the noise introduced by the instrument itself [44].
- Apply Filtering: Use software-based band-pass filters to remove predictable, high-frequency noise that falls outside the bandwidth of your signal of interest [44].
Issue 4: Optimizing Multiple Conflicting Experimental Factors
Problem: An experiment or process has several input factors (e.g., temperature, pH, concentration) that interact and conflict in their effects on multiple output criteria (e.g., yield, cost, purity). Solution:
- Apply the Simplex Method: This is an iterative search optimization method that is effective for navigating multi-parameter spaces, especially when the mathematical relationship between inputs and outputs is complex or unknown [45].
- Formulate a Multi-Criteria Problem: Define your input factors and the multiple output criteria you wish to optimize. Involve domain experts or a decision maker (DM) to fuzzily evaluate criteria that are difficult to quantify physically (e.g., "product quality") [45].
- Find a Pareto-Optimal Solution: Use a modified simplex method combined with principles like Pareto optimality to identify a set of solutions where no single criterion can be improved without worsening another. This allows you to select the best compromise solution for your specific needs [45].
Data Presentation
Table 1: Quantitative Relationships in a Model Genetically Encoded Oscillator (GEO)
This table summarizes how different expression strategies and noise types affect the waveform of a synthetic protein oscillator, based on empirical characterization [40].
| Experimental Factor | Impact on Oscillation Frequency | Impact on Oscillation Amplitude | Primary Noise Type Affected |
|---|---|---|---|
| Absolute ATPase Abundance | Minimal direct impact | Strong positive correlation; amplitude scales with ATPase level | Extrinsic Noise |
| Activator:ATPase Ratio | Strong positive correlation; frequency increases with ratio | Bell-shaped response; peaks within functional bandwidth | Intrinsic Noise |
| Genomic Integration (Single Copy) | High stability; low population variance | Stable, but locked to a specific level | Attenuates Extrinsic Noise |
| Multi-Copy Plasmid Expression | High population variance | High population variance | Amplifies Both Intrinsic & Extrinsic Noise |
| Functional Bandwidth Boundaries | Oscillations cease outside min/max ratio | Amplitude drops to background levels outside boundaries | N/A |
Table 2: Research Reagent Solutions for Noise Management
This table lists key reagents, components, and computational tools used to study and manage biological noise.
| Reagent / Tool Name | Type / Category | Primary Function in Noise Management |
|---|---|---|
| Dual-Fluorescence Reporter System | Experimental Molecular Tool | Empirically distinguishes between intrinsic and extrinsic noise in gene expression [38] [41]. |
| Tef1 Constitutive Promoter | DNA Part (Yeast) | Provides strong, stable expression; used to minimize population-level variation in genomically integrated circuits [40]. |
| scDist | Computational Tool (Bioinformatics) | Detects transcriptomic differences in single-cell data while minimizing false positives from individual and cohort variation [43]. |
| MMIDAS (Mixture Model Inference with Discrete-coupled Autoencoders) | Computational Tool (Bioinformatics) | Learns discrete cell clusters and continuous, cell-type-specific variability from unimodal and multimodal single-cell datasets [43]. |
| MinDE-family ATPase/Activator Pairs | Protein Components (Synthetic Biology) | Core components for constructing genetically encoded oscillators (GEOs); model system for studying noise in protein circuits [40]. |
| Constrained Disorder Principle (CDP) | Theoretical Framework | Guides the design of regimens (e.g., varied drug administration) that use controlled noise to improve system function and overcome tolerance [43]. |
Experimental Protocols
Protocol 1: Mapping a Phase Portrait for a Synthetic Biological Oscillator
Objective: To experimentally define the relationship between component expression levels and the dynamic output of a genetically encoded oscillator (GEO) [40]. Materials:
- Budding yeast (Saccharomyces cerevisiae) strain.
- Expression plasmids for GEO components (e.g., MinD ATPase and MinE Activator, fluorescently tagged) on both high- and low-copy plasmids.
- Timelapse fluorescence microscopy setup.
- Image analysis software with Fast Fourier Transform (FFT) capability.
Methodology:
- Generate Diverse Expression Population: Transform yeast with pairwise combinations of ATPase and Activator plasmids to create a population of cells with a wide range of expression level combinations.
- Image Live Cells: Perform timelapse fluorescence microscopy on the population. Capture both fluorescence images (to quantify component levels) and phase-contrast images.
- Quantify Single-Cell Expression and Dynamics: For each cell, use background-corrected fluorescence intensity as a proxy for protein abundance. Use FFT on the time-lapse signal from each pixel or cellular region to determine the oscillation status (on/off), frequency (mHz), and amplitude.
- Construct Phase Portraits: Aggregate data from thousands of cells. Create 2D plots with ATPase and Activator levels on the axes, and color-code the data points to represent oscillation frequency, amplitude, or state. Use machine learning classifiers (e.g., logistic regression) to precisely define the boundaries of the oscillatory regime.
Objective: To empirically find the excitation voltage that maximizes the Signal-to-Noise Ratio (SNR) for a strain gauge without causing instability due to self-heating [44]. Materials:
- Strain gauge installed on the test material.
- Strain gauge amplifier/conditioner with adjustable excitation voltage.
- Data acquisition system.
- Thermal camera (optional, to monitor gauge temperature).
Methodology:
- Initial Setup: With no load applied to the strain gauge, zero the measurement channel.
- Progressive Voltage Increase: Gradually increase the bridge excitation voltage in small increments.
- Monitor Zero-Point Stability: At each voltage level, observe the zero reading of the channel over a period of time. Note the voltage level at which the zero reading becomes unstable (shows significant drift).
- Establish Optimal Voltage: Reduce the excitation voltage to the highest level that maintains a stable zero reading. This is the optimal voltage for your specific installation. For greater confidence, perform this test at the highest environmental temperature the gauge will experience during actual measurements.
Mandatory Visualization
Diagram 1: Noise Propagation in a Synthetic Genetic Oscillator
Title: How Intrinsic and Extrinsic Noise Affect Oscillator Outputs
Diagram 2: Sequential Simplex Optimization of Multiple Factors
Title: Workflow for Multi-Criteria Simplex Optimization
The sequential simplex method is an evolutionary operation (EVOP) technique used for optimizing multiple experimental factors. It is a direct search algorithm that operates by moving a geometric figure (a simplex) through the experimental space based on the results of previous experiments. For an optimization problem with k factors, the simplex typically consists of k+1 vertices. At each step, the algorithm reflects the worst-performing vertex through the centroid of the opposite face, testing new conditions while progressively moving toward more optimal regions. Unlike the Nelder-Mead variable simplex procedure used for numerical optimization, the basic simplex method for process improvement uses fixed step sizes to minimize the risk of producing non-conforming products during experimentation [46].
Troubleshooting Guides and FAQs
FAQ 1: How do I determine the appropriate initial step size for my experiment?
Answer: The initial step size (factorstep dxi) represents the distance between your initial measurement points in each factor dimension. Selection requires balancing two competing concerns:
- Too large: High risk of producing non-conforming products and moving into undesirable operating regions.
- Too small: The signal-to-noise ratio (SNR) may be insufficient to pinpoint the direction of the optimum against background process variation [46].
A practical approach is to start with a step size that represents a small, manageable perturbation from your current best-known operating conditions—typically 5-15% of your factor's operational range. This is small enough to keep the process within acceptable specifications but large enough to generate a detectable effect over process noise.
FAQ 2: Why is my simplex process failing to converge to a better optimum?
Answer: Failure to converge can stem from several issues:
- Excessive Noise: If the process noise is high relative to the effect of factor changes (low SNR), the algorithm cannot reliably determine the correct direction to move. Visually, the response becomes overly obscured, making progression difficult [46].
- Oversized Simplex: A step size that is too large may cause the simplex to jump over the optimal region entirely.
- Ridge Following: The simplex can become stuck moving slowly along a ridge in the response surface.
Troubleshooting Protocol:
- Verify SNR: Conduct a preliminary study to estimate your process variability. Ensure that the expected improvement from a single simplex step is large compared to the standard deviation of your response measurement.
- Reduce Step Size: Systematically reduce the reflection step size. While this may slow convergence, it improves stability in noisy environments.
- Implement a "Soft Restart": If the simplex has been progressing slowly for many iterations, re-initialize it around the current best vertex with a potentially smaller step size to re-focus the search.
FAQ 3: How can I accelerate convergence without increasing the risk of failure?
Answer: Accelerating convergence while maintaining safety is a key challenge.
- Use a Hybrid Approach: Once the simplex has moved decisively away from the initial region and into a promising area, consider a controlled increase in step size for a single reflection. This should only be done when recent moves have consistently shown improvement, indicating a strong gradient.
- Leverage Process Knowledge: If you have prior mechanistic knowledge or a preliminary model, use it to bias the initial simplex orientation or to take a larger step in a direction known to be safe and impactful.
- Implement a Convergence Boosting Scheme: For the sequence of results obtained, apply a simple smoothing technique. For successive values f(k, x), compute a smoothed version:
g(k, x, 1) = 0.5 * f(k, x) + 0.5 * f(k-1, x). This can help dampen noise and clarify the underlying trend, allowing for more confident step selection [47].
FAQ 4: What is the practical difference between using Simplex and EVOP for process improvement?
Answer: Both methods use small perturbations for online optimization, but their operational logic differs. The table below summarizes the key distinctions relevant to a researcher.
Table 1: Comparison of Simplex and EVOP Methods for Process Improvement
| Feature | Sequential Simplex Method | Evolutionary Operation (EVOP) |
|---|---|---|
| Core Mechanism | Moves a geometric simplex; replaces the worst vertex per iteration [46] | Uses a pre-designed factorial array (e.g., 2^2) centered at the current condition [46] |
| Experiments per Step | Adds only one new point per iteration [46] | Requires a full cycle of experiments (e.g., 4 points + center) per phase [46] |
| Information Usage | Uses only ranking of vertices; less sensitive to absolute scaling | Uses a linear model to estimate gradient; requires more precise measurements |
| Strengths | Simpler calculations, minimal experiments per step, efficient movement [46] | Can handle qualitative factors, builds a local model [46] |
| Weaknesses | Prone to getting lost in very noisy environments [46] | Becomes prohibitively expensive with many factors [46] |
Quantitative Data and Experimental Protocols
The following table summarizes findings from a simulation study comparing Simplex and EVOP under different experimental conditions. This data can guide your choice of method and step size.
Table 2: Performance of Simplex and EVOP Under Varying Conditions [46]
| Condition | Performance Metric | Simplex Method | EVOP Method |
|---|---|---|---|
| Low Dimensionality (k=2-4) | Steps to Converge | Fastest | Slower |
| High Dimensionality (k>5) | Steps to Converge | Performance degrades | More reliable |
| High Noise (Low SNR) | Success Rate | Low | Higher, more robust |
| Small Factorstep (dxi) | Convergence Speed | Slow, can stall | More consistent progress |
| Large Factorstep (dxi) | Risk of Non-Conforming Product | High | High |
Experimental Protocol: Setting Up an Initial Simplex
Objective: To correctly initialize the sequential simplex procedure for a new optimization problem.
Materials:
- The process or system to be optimized.
- Identified
kcontinuous factors (Xâ‚, Xâ‚‚, ..., Xâ‚–) to adjust. - A single quantifiable response variable (Y) to maximize or minimize.
- Defined operational ranges and constraints for all factors to avoid non-conforming product.
Procedure:
- Define the Base Point (Vertex 0): Choose a set of factor levels known to produce acceptable product quality. This is your starting point, Vâ‚€ = (xâ‚â‚€, xâ‚‚â‚€, ..., xâ‚–â‚€).
- Select Step Sizes (dxi): For each factor
i, choose a step sizedxáµ¢. This is a critical choice that balances speed and safety, as detailed in FAQ 1. - Calculate Initial Simplex Vertices: The remaining
kvertices are generated from the base point using the step sizes.- Vertex 1:
Vâ‚ = (xâ‚â‚€ + dxâ‚, xâ‚‚â‚€, ..., xâ‚–â‚€) - Vertex 2:
Vâ‚‚ = (xâ‚â‚€, xâ‚‚â‚€ + dxâ‚‚, ..., xâ‚–â‚€) - ...
- Vertex k:
Vâ‚– = (xâ‚â‚€, xâ‚‚â‚€, ..., xâ‚–â‚€ + dxâ‚–)
- Vertex 1:
- Run Experiments: Conduct the process at each of the
k+1vertices (Vâ‚€, Vâ‚, ..., Vâ‚–) and measure the responseY. - Begin Iteration: Identify the vertex with the worst response (for minimization) and proceed with the reflection step to generate a new vertex.
Workflow Visualization
The following diagram illustrates the logical workflow of the sequential simplex method, integrating step size decisions and quality checks.
Diagram 1: Sequential Simplex Workflow with Quality Gate
The Scientist's Toolkit: Research Reagent Solutions
This table outlines key conceptual "reagents" essential for designing and executing a sequential simplex optimization study.
Table 3: Essential Components for a Sequential Simplex Experiment
| Item | Function/Explanation | Example/Consideration |
|---|---|---|
| Defined Factors (Xâ‚...Xâ‚–) | The input variables to be adjusted to improve the process. | Temperature, pressure, catalyst concentration, reaction time. |
| Quantified Response (Y) | The single output metric used to evaluate performance. | Yield, purity, particle size, production rate. Must be measurable with low noise. |
| Base Point (Vâ‚€) | The set of initial factor levels representing a known, acceptable process condition. | Prevents starting from a poor or risky region, anchoring the search in a "safe" zone. |
| Step Size (dxáµ¢) | The magnitude of the initial perturbation for each factor. | The critical parameter for balancing convergence speed and risk of failure. |
| Stopping Rule | Pre-defined criteria to halt experimentation. | e.g., % improvement < threshold for N consecutive steps, or maximum iterations reached. |
| Constraint Definitions | Operational boundaries that define a "non-conforming product." | e.g., Purity must be >95%, a byproduct concentration must be <0.1%. Used in the quality check. |
Frequently Asked Questions (FAQs)
FAQ 1: What is the most common cause of a simplex appearing to "stick" or circle in a small area of the experimental domain? This behavior often indicates the simplex is operating near a local optimum or on a flat response surface. To continue progression, apply a "change of direction" rule: instead of rejecting the vertex with the worst response, reject the vertex with the second-worst response and reflect it across the line defined by the two remaining vertices. This changes the simplex's progression axis and helps explore new regions. If circling persists, consider expanding the simplex size to escape the local area [9].
FAQ 2: How do I handle experimental factors that have strict, immovable boundaries? When the reflection of a vertex lands outside a feasible experimental boundary, you must create a new vertex at the boundary itself. However, simply placing it on the boundary is often insufficient. Instead, assign an exceptionally poor performance value to this boundary point in your objective function. This ensures the simplex algorithm automatically rejects this vertex on the next iteration and moves away from the infeasible region, effectively treating the boundary as a constraint that should not be explored [9].
FAQ 3: Why does my sequential simplex method perform poorly when my experimental data has high noise levels? The simplex method relies on clear differences between vertex performances to determine direction. Noisy data obscures these differences. Implement a "weighted centroid" calculation that gives higher importance to vertices that have consistently shown better performance over multiple iterations. Furthermore, consider increasing the step size of your reflections slightly beyond the standard reflection coefficient to ensure the simplex moves decisively away from areas where signal-to-noise ratio is low [48].
FAQ 4: What is the critical difference between the basic simplex and modified simplex methods for handling constraints? The basic simplex method maintains a constant size and only reflects vertices, which can limit its ability to navigate around constraints. The modified simplex method (Nelder-Mead) can expand, contract, or reflect, giving it flexibility. When encountering constraints, the modified method can contract to carefully navigate along constraint boundaries or expand to move rapidly away from infeasible regions once they are identified [9] [48].
Troubleshooting Guides
Problem: Simplex Oscillation Between Two States
Symptoms: The algorithm alternates between two similar simplex configurations without meaningful progress.
Resolution Steps:
- Identify: Monitor for two consecutive vertices being reflected back and forth.
- Apply Rule 2: Reject the vertex with the second-worst response instead of the worst.
- Verify: Check that the new direction produces a meaningful change in the response value.
- Prevent: Implement a memory of recently visited vertices to avoid immediate backtracking [9].
Problem: Vertex Lands in an Infeasible Experimental Region
Symptoms: A calculated vertex requires factor levels outside safe or possible operating conditions.
Resolution Steps:
- Abort: Do not run the experiment with infeasible parameters.
- Assign Penalty: Assign a mathematically poor value (e.g., zero yield, or negative infinity for maximization) to this vertex in your objective function.
- Contract: Perform a contraction step toward the best vertex instead of a reflection.
- Resume: Continue with the new, contracted simplex away from the constraint [9].
Problem: Degenerate Simplex (Loss of Dimensionality)
Symptoms: The simplex becomes excessively flat or elongated, reducing search efficiency.
Resolution Steps:
- Detect: Calculate the volume of the simplex; a near-zero volume indicates degeneracy.
- Reset: If detected, restart a new simplex in a different region of the experimental domain, using knowledge gained from previous iterations.
- Prevent: Regularly check the condition number of the simplex vectors; if it becomes too high, introduce a small random perturbation to non-best vertices to restore dimensionality [49].
Experimental Protocol: Implementing a Constraint-Handling Simplex for Chemical Synthesis
This protocol details the methodology for optimizing a chemical synthesis using a microreactor system with inline monitoring, based on published research [48].
Objective: To maximize the yield of n-benzylidenebenzylamine (3) from the condensation of benzaldehyde (1) and benzylamine (2) by optimizing residence time and temperature, while respecting equipment and safety constraints.
Research Reagent Solutions
| Item Name | Function/Brief Explanation |
|---|---|
| Benzaldehyde (1) | Primary reactant substrate (ReagentPlus, 99%) [48]. |
| Benzylamine (2) | Primary reactant substrate (ReagentPlus, 99%) [48]. |
| Methanol | Reaction solvent (for synthesis, >99%) [48]. |
| Stainless Steel Capillaries | Microreactor components (0.5 mm & 0.75 mm id) for continuous flow synthesis [48]. |
| Syringe Pumps (SyrDos2) | For precise, continuous dosage of starting materials [48]. |
| Inline FT-IR Spectrometer | For real-time reaction monitoring via characteristic IR bands (e.g., 1680-1720 cmâ»Â¹ for benzaldehyde) [48]. |
| Laboratory Automation System | Controls temperature and flow rates; integrates pumps, analytics, and optimisation software [48]. |
| MATLAB Software | Executes the experimental sequence, optimisation algorithm, and calculates the objective function [48]. |
Methodology
- Initialization: Prepare a stock solution of benzaldehyde (1) and benzylamine (2) in methanol at initial concentrations of 4 mol Lâ»Â¹ each [48].
- Setup Configuration: Assemble the microreactor system consisting of two connected coiled capillaries (total volume 1.87 mL). Connect the syringe pumps, thermostats, and the inline FT-IR spectrometer to the automation system [48].
- Define Constraints: Set hard boundaries for factors:
- Temperature: 20°C to 80°C (based on solvent boiling point and equipment limits).
- Residence Time: 0.5 to 6 minutes (based on reactor volume and pump flow rate capabilities).
- Initial Simplex: Define a starting simplex (triangle) within the feasible region of temperature and residence time.
- Automated Optimization Loop:
- The automation system executes experiments at each vertex of the current simplex.
- The inline FT-IR spectrometer monitors the reaction in real-time, calculating the conversion of benzaldehyde and the yield of the imine product based on pre-determined calibration curves [48].
- The yield is used as the objective function value for each vertex.
- The MATLAB script implements the modified simplex algorithm:
- Reflect the worst vertex.
- If the new vertex violates constraints, assign a yield of 0% and contract the simplex.
- If the new vertex is feasible and shows improved yield, expand.
- New setpoints for temperature and flow rates (affecting residence time) are sent to the automation system.
- The loop continues until the simplex converges to an optimum or a maximum number of iterations is reached.
Workflow Diagram
Troubleshooting Guides
Guide 1: Diagnosing and Resolving Simplex Oscillation
Problem Identification: Simplex oscillation occurs when the algorithm cycles between several vertices without making progress toward the optimum. This frequently happens in degenerate problems where multiple vertices correspond to the same objective function value.
Diagnostic Steps:
- Monitor Vertex Changes: Track the basic variables and their values through iterations. If a sequence of bases repeats, oscillation is confirmed.
- Check Objective Stagnation: If the objective function value remains constant for multiple iterations despite changes in the basis, this indicates degeneracy.
- Analyze Pivot Selections: Note if the entering and leaving variables are simply swapping roles in a cyclical pattern.
Resolution Protocol:
- Apply Bland's Rule: To prevent cycling, implement a deterministic pivot selection rule. Choose the entering variable with the smallest index among those with negative reduced costs, and similarly, select the leaving variable with the smallest index in case of ties in the minimum ratio test [50].
- Perturbation Method: Add infinitesimal constants (ε, ε², ..., εáµ) to the right-hand side (RHS) of constraints. This technique breaks degeneracy by ensuring that no two vertices have the exact same solution values, preventing the simplex from cycling between them.
- Lexicographic Ordering: Use a lexicographic method for selecting the leaving variable to ensure a unique pivot path and avoid loops.
Guide 2: Addressing False Convergence in Modified Simplex Methods
Problem Identification: False convergence occurs when the simplex algorithm stagnates at a point that is not the true optimum, often due to the simplex becoming overly small or distorted on the response surface.
Diagnostic Steps:
- Measure Simplex Size: Calculate the volume or the lengths of the edges of the current simplex. A significant reduction in size suggests the simplex is collapsing.
- Check Vertex Responses: Compare the objective function values at all vertices. If they are very similar but suboptimal, false convergence is likely.
- Assess Simplex Degeneracy: Evaluate the shape of the simplex by calculating the angles between its edges. Very small angles indicate a degenerate, "flat" simplex that cannot move effectively [34].
Resolution Protocol:
- Implement Expansion Steps: Use a larger expansion coefficient (e.g., 2.2–2.5) to allow the simplex to take larger steps and explore a broader area of the response surface [34].
- Constraint on Degeneracy: Introduce a computational constraint that monitors the angles within the simplex. If the simplex becomes too degenerate, trigger a reset or translation procedure to restore a healthier shape [34].
- Simplex Translation: Upon repeated failed contractions, translate the entire simplex while retaining its size and shape. This moves the entire search region to escape the area of stagnation [34].
- Boundary Handling: If a vertex moves outside a variable's feasible boundary, correct it back to the boundary instead of simply assigning it an unfavorable value. This improves performance for optima located on constraints [34].
Frequently Asked Questions (FAQs)
FAQ 1: What are the most common signs that my simplex optimization is oscillating or has converged to a false optimum?
The key indicators are:
- Oscillation: The algorithm cycles through a repeating sequence of bases or vertices without improving the objective function.
- False Convergence: The simplex size becomes infinitesimally small, or the objective function values at all vertices converge to a similar, yet suboptimal, value. The algorithm stops without satisfying the true optimality conditions.
FAQ 2: Which specific simplex method modifications are most effective for preventing these convergence issues?
Research on test functions indicates that the most reliable method combines several modifications [34]:
- Type B Method: A specific modification for handling expansion and contractions.
- Degeneracy Constraint: A mechanism to control the shape of the simplex and prevent it from becoming degenerate.
- Translation: Moving the entire simplex after repeated failed contractions.
- Boundary Correction: Correcting vertices that fall outside constraints back to the boundary instead of penalizing them.
FAQ 3: How do I handle variable boundaries to avoid convergence problems?
When a vertex is located outside a variable's feasible boundary, the most effective strategy is to correct the vertex by moving it back onto the boundary. This approach has been shown to increase the speed and reliability of convergence compared to simply assigning the vertex an unfavorable response value [34].
FAQ 4: Are there proven parameter values for reflection, contraction, and expansion that improve convergence?
Yes, parameter studies have found that a reflection coefficient of 1.0 and a contraction coefficient of 0.5 are nearly optimal for many functions. For the expansion coefficient, a value larger than 2.0, specifically in the range of 2.2 to 2.5, is generally more appropriate and enables the simplex to search a larger area effectively [34].
Research Reagent Solutions
The following table details key computational strategies and their roles in troubleshooting simplex convergence issues.
| Reagent/Tool | Function in Optimization |
|---|---|
| Bland's Rule | An anti-cycling algorithm that ensures convergence by providing a deterministic rule for pivot selection [50]. |
| Expansion Coefficient (γ > 2.0) | A parameter that controls the size of the expansion step, allowing the simplex to move faster in favorable directions [34]. |
| Degeneracy Constraint | A computational check that prevents the simplex from becoming overly flat and numerically unstable, thus maintaining its search efficiency [34]. |
| Simplex Translation | A recovery procedure that moves the entire simplex to a new location to escape regions of repeated failed contractions [34]. |
Experimental Protocols & Data
Protocol 1: Evaluating Simplex Method Performance
Objective: To compare the convergence reliability and speed of different simplex variants on standard test functions.
Methodology:
- Select Test Functions: Use a set of established multi-parameter test functions (e.g., Powell's function, Wood's function) with known optima and complex response surfaces [34].
- Configure Algorithms: Implement the Basic Simplex Method (BSM), Modified Simplex Method (MSM), and improved variants (e.g., Type B with degeneracy constraint and translation).
- Run Optimizations: For each method and function, run multiple optimizations from different starting points.
- Collect Data: Record the number of iterations (or function evaluations) until convergence and note instances of non-convergence (failure).
Expected Outcome: A comparison table showing the success rate and average number of evaluations for each method, highlighting the robustness of the improved variants.
Quantitative Performance Data
The table below summarizes findings from a study that evaluated different simplex method modifications across nine test functions [34].
| Simplex Method Modification | Key Performance Finding |
|---|---|
| Basic Simplex Method (BSM) | Prone to failure on complex surfaces and near variable boundaries. |
| Modified Simplex Method (MSM) | Improved efficiency over BSM but can still suffer from degeneracy and false convergence. |
| Type B with Translation & Degeneracy Constraint | Most reliable method for finding the optimum region across diverse test functions. |
| Expansion Coefficient (γ = 2.2 to 2.5) | Increased search area and reduced the number of evaluations required for convergence. |
| Boundary Correction (vs. Penalty) | Increased the speed and reliability of convergence for optima on variable boundaries. |
Workflow and Relationship Visualizations
Diagram 1: Convergence problem troubleshooting workflow.
Troubleshooting Guides
FAQ 1: Why would I combine the Simplex method with other optimization algorithms, and what are the primary benefits?
Answer: Combining the Simplex method with other algorithms aims to enhance performance by leveraging the strengths of different approaches. The primary benefits include:
- Reduced Iteration Count: Hybrid methods can find a better starting point for the Simplex algorithm, allowing it to reach the optimal solution in fewer iterations [51]. This avoids the Simplex method's potentially long path along the boundaries of the feasible region.
- Global Optimization Capabilities: The standard Nelder-Mead Simplex method is a local optimizer. When hybridized with global search techniques like Inductive Search, the algorithm gains the ability to locate global minima in complex, multi-modal problem landscapes, which is crucial for challenging applications like protein folding [52].
- Improved Computational Efficiency: By reducing the number of iterations required, the total running time to find the optimal solution can be significantly decreased, as demonstrated in tests on standard linear programming (LP) problem sets [51].
FAQ 2: My hybrid algorithm is not converging, or it converges to a local minimum instead of a global one. What could be wrong?
Answer: Failure to converge or convergence to a local optimum is a common issue in optimization. Below is a structured guide to troubleshoot this problem.
| Problem Area | Specific Issue | Diagnostic Steps | Proposed Solution |
|---|---|---|---|
| Global Search Phase | The global search is insufficiently exploring the parameter space. | Check the diversity of points sampled in the initial phases. | Increase the scope of the global search component (e.g., Brent's method in a Hybrid Global Optimizer) or incorporate adaptive random search to ensure a thorough exploration [52]. |
| Transition to Local Search | The algorithm transitions to the Simplex method from a poor starting point. | Log the objective function value at the transition point. | Implement a more robust method to find an advanced starting point. For LP problems, using an interior search direction to find an improved basic feasible solution can be more effective [51]. |
| Algorithm Parameters | Parameters (e.g., for reflection, expansion in Nelder-Mead) are poorly tuned for your specific problem. | Perform a sensitivity analysis on the algorithm's key parameters. | Systematically tune parameters. For instance, in a hybrid-LP method, parameters like β (in the range 0.7–1.0) are critical and may need optimization for different problem types [51]. |
| Problem Formulation | The problem is non-convex or has noisy gradients. | Review the problem's mathematical properties. | Ensure the hybrid method is appropriate. A hybrid like the one involving Simplex and Inductive Search is specifically designed for difficult, non-linear global optimization problems [52]. |
FAQ 3: The optimization process is computationally expensive and slow. How can I improve its performance?
Answer: Performance bottlenecks can occur in different stages of the hybrid algorithm. This guide helps identify and address them.
| Problem Area | Specific Issue | Diagnostic Steps | Proposed Solution |
|---|---|---|---|
| Function Evaluations | The objective function itself is computationally expensive to calculate. | Profile your code to confirm the time is spent in the function evaluation. | Optimize the function code or use surrogate models. The hybrid method itself aims to reduce the number of expensive iterations [52]. |
| Global Search Overhead | The global search phase is taking too long before handing over to the efficient Simplex. | Compare the time spent in the global phase versus the local Simplex phase. | Adjust the termination criteria for the global search. The goal is to find a "good enough" starting point efficiently, not the perfect one [51]. |
| Simplex Iterations | The Simplex method is still taking many iterations after the hybrid handoff. | Record the number of Simplex iterations from the hybrid starting point versus a standard starting point. | The hybrid starting point should be significantly improved. If not, revisit the global search strategy. Research indicates that a well-designed hybrid-LP can reduce both iterations and running time [51]. |
| Implementation Code | The algorithm is implemented in an interpreted language without optimization. | Check if the core computational loops can be optimized. | Use optimized libraries or consider a compiled language for critical sections. The literature notes that a simple implementation in MATLAB can show promise, but an optimized implementation would yield better performance [51]. |
Experimental Protocols
Protocol 1: Implementing a Hybrid Global Optimizer with Simplex and Inductive Search
This protocol is based on the hybrid algorithm combining the Nelder-Mead Simplex method with Inductive Search for global optimization, suitable for complex problems like protein folding energy minimization [52].
1. Objective: To minimize a non-linear, potentially multi-modal function where finding the global minimum is critical.
2. Materials and Reagent Solutions:
- Software Environment: A computational mathematics environment (e.g., MATLAB, Python with SciPy).
- Core Algorithms: Code implementations of:
- Nelder-Mead Simplex method (local search)
- Brent's method for line minimization (global search)
- Inductive Search routines (global search)
3. Methodology:
- Step 1: Global Search Phase (Inductive Search & Brent's Method). Use Inductive Search to strategically explore the parameter space. Within promising regions, employ Brent's method for efficient line minimizations and the Simplex method for plane minimizations to locate a candidate solution basin [52].
- Step 2: Solution Handoff. The best point(s) found in the global search phase are used as the starting point(s) for the local search phase.
- Step 3: Local Search Phase (Nelder-Mead Simplex). Initialize the Nelder-Mead Simplex algorithm around the promising starting point. Execute the Simplex method with its standard operations (reflection, expansion, contraction) to converge to a high-precision local minimum [52].
- Step 4: Validation. Compare the result against known global optima (for test functions) or run the algorithm multiple times with different initial global search seeds to check for consistency.
The following workflow diagram illustrates the logical relationship and data flow between the components of this hybrid optimizer:
Protocol 2: Implementing a Hybrid-LP Method for an Improved Simplex Starting Point
This protocol is for linear programming (LP) problems and details a two-step method where an interior search finds an improved starting point for the Simplex method [51].
1. Objective: To solve an LP problem of the form Maximize (z = c^Tx) subject to (Ax = b, x \geq 0), with reduced iterations and run-time.
2. Materials and Reagent Solutions:
- Software Environment: A platform with linear programming capabilities (e.g., MATLAB, Python with PuLP/SciPy).
- Core Algorithms: Code for:
- Standard Simplex method
- Hybrid-LP interior direction search
3. Methodology:
- Step 1: Problem Setup. Convert the LP problem into standard form.
- Step 2: Interior Direction Search. From an initial feasible point, compute a search direction that moves through the interior of the feasible region, avoiding the boundary. This direction is derived from the reduced gradient to improve the objective function value [51].
- Step 3: Pivot to a Basic Feasible Solution (BFS). Move along the interior direction until a constraint is encountered. Perform a pivot operation to land on an improved BFS. This is the advanced starting point for the next phase [51].
- Step 4: Simplex Phase. Use the standard Simplex method, starting from the improved BFS obtained in Step 3. Continue with standard Simplex pivoting until the optimal solution is reached [51].
The workflow for this LP-specific hybrid method is shown below:
The Scientist's Toolkit: Research Reagent Solutions
The following table details key computational components and their roles in implementing hybrid simplex algorithms.
| Item Name | Function in Experiment | Specific Application Note |
|---|---|---|
| Nelder-Mead Simplex | A direct search method for finding a local minimum of an objective function in a multi-dimensional space. | Serves as the efficient local refinement engine in the hybrid structure. It is robust but can get stuck in local minima [52]. |
| Inductive Search | A global search technique that strategically explores the parameter space to identify promising regions containing the global minimum. | Used in the initial phase to guide the search away from poor local minima and towards the global basin [52]. |
| Brent's Method | A root-finding and minimization algorithm combining bisection, secant, and inverse quadratic interpolation. | Employed within the hybrid for highly efficient line minimizations along promising search directions [52]. |
| Interior Search Direction (Hybrid-LP) | A search direction derived from the reduced gradient that moves through the interior of the feasible region in an LP problem. | Aims to find a superior starting point for the Simplex method, reducing the number of subsequent boundary pivots [51]. |
| Reduced Gradient | The gradient of the objective function projected onto the null space of the active constraints. | Fundamental for calculating feasible improving directions in constrained problems, such as in the Hybrid-LP method [51]. |
Benchmarking Sequential Simplex: Performance Validation and Method Comparison in Research Contexts
Troubleshooting Guides
Guide 1: Diagnosing Unacceptable Algorithm Runtimes
Problem: Your computational experiment is taking too long to produce results, causing significant delays in your research timeline. Explanation: This is a classic symptom of encountering exponential time complexity. When the number of experimental factors increases, algorithms with exponential runtimes can become infeasible.
Diagnostic Steps:
- Identify Input Size: Determine the parameter
nthat scales in your experiment (e.g., number of factors, data points, or components). - Benchmark Runtime: Measure the execution time for progressively larger input sizes (
n,n+1,n+2). - Analyze Growth:
Solution:
- For exponential problems, reframe the problem to use a heuristic or approximation algorithm.
- Where possible, leverage algorithms known to have polynomial time complexity for the specific sub-problem you are solving.
Guide 2: Differentiating Complexity Classes in Experimental Data
Problem: You have plotted the runtime of your procedure against the input size but are unsure how to classify the growth.
Diagnostic Steps:
- Plot on a Log-Linear Scale: Plot the input size
nagainst the logarithm of the runtime (log(T(n))). - Interpret the Plot:
- A straight line suggests polynomial growth, O(náµ).
- A curve that itself increases exponentially suggests exponential growth, O(câ¿) [53].
Solution: Use this classification to select the appropriate optimization strategy in your sequential simplex method. Polynomial growth is generally more manageable.
Frequently Asked Questions (FAQs)
Q1: What is the fundamental difference between polynomial and exponential time complexity?
The core difference lies in how the resource requirements grow as the input size n increases. Polynomial complexity (O(náµ)) grows at a rate proportional to n raised to a constant power k, leading to manageable and predictable growth [53]. In contrast, exponential complexity (O(câ¿)) grows at a rate proportional to a constant c raised to the power of n, resulting in rapid, often unmanageable growth that quickly makes problems intractable for even moderately sized inputs [53].
Q2: Why should I care about this distinction in experimental optimization and drug development?
Understanding this distinction is crucial for project feasibility and resource allocation. Algorithms with polynomial time complexity are generally considered efficient and scalable, making them suitable for analyzing larger datasets or optimizing processes with more factors [53]. Algorithms with exponential complexity are often intractable for large inputs; encountering one in your research is a signal that you may need to use approximations, heuristics, or focus on smaller sub-problems to get results in a reasonable time [53].
Q3: Can you provide concrete examples of algorithms in each category?
Certainly. Common examples of polynomial time algorithms include:
Common examples of exponential time problems include:
- The Traveling Salesman Problem (solved via brute-force search) [54]
- The Subset Sum Problem (O(2â¿)) [53]
- The Knapsack Problem (O(2â¿)) [53]
Q4: How does the Sequential Simplex Method relate to computational complexity?
The Sequential Simplex Method is an optimization algorithm itself, used to find the best experimental conditions by navigating a multi-dimensional space [11]. Its efficiency can be analyzed in terms of time complexity. Furthermore, it is often employed as a heuristic to find good solutions for complex problems that may otherwise require algorithms with much worse (e.g., exponential) time complexity, thereby making optimization in multi-factor experiments computationally feasible [12].
Data Presentation: Complexity Comparison
The following table summarizes the key differences between polynomial and exponential complexities, which is essential for diagnosing computational bottlenecks in research.
Table 1: Characteristic Differences Between Polynomial and Exponential Complexities [53]
| Aspect | Polynomial Complexity | Exponential Complexity |
|---|---|---|
| Definition | O(náµ) for some constant k | O(câ¿) for some constant c>1 |
| Growth Rate | Grows at a rate proportional to náµ | Grows at a rate proportional to câ¿ |
| Feasibility | Manageable and predictable growth; feasible for larger inputs | Rapid, unmanageable growth; quickly becomes infeasible |
| Scalability | Highly scalable | Poor scalability |
| Typical Use Cases | Suitable for problems with larger input sizes | Used for NP-hard problems or only with small input sizes |
| Example Algorithms | Linear Search (O(n)), Bubble Sort (O(n²)) | Subset Sum (O(2â¿)), Brute-Force TSP (O(n!)) |
Experimental Protocol: Classifying Algorithm Growth
Objective: To empirically determine whether an unknown algorithm or experimental procedure exhibits polynomial or exponential time complexity.
Materials:
- The algorithm or procedure to be tested.
- A computer system with consistent performance.
- Data generation tools to create input sets of varying sizes
n. - A timing mechanism (e.g., system clock, profiling software).
Methodology:
- Input Generation: Prepare a series of input sets, with sizes
nranging from small (e.g., 5, 10) to the largest feasible (e.g., 100, 500, 1000). Ensure the input structure is consistent across sizes. - Runtime Measurement: For each input size
n, execute the algorithm at least three times and record the average runtimeT(n). - Data Transformation: Create a new dataset where you take the logarithm of the runtime,
log(T(n)), for each input sizen. - Data Analysis: Plot two graphs:
- Graph A: Input Size (
n) vs. Runtime (T(n)). - Graph B: Input Size (
n) vs. Log Runtime (log(T(n))).
- Graph A: Input Size (
Interpretation:
- If Graph B (log-linear) approximates a straight line, the algorithm likely has polynomial time complexity.
- If Graph A shows a curve that increases extremely rapidly and Graph B shows a steep, non-linear curve, the algorithm likely has exponential time complexity.
Mandatory Visualizations
Diagram 1: Complexity Growth Comparison
Diagram 2: Problem Complexity Decision Tree
The Scientist's Toolkit: Research Reagent Solutions
Table 2: Essential Computational Tools for Complexity Analysis
| Tool Name | Function & Purpose | Relevance to Research |
|---|---|---|
| Sequential Simplex Method | An optimization algorithm that uses a geometric figure to navigate a multi-dimensional parameter space towards an optimum [11]. | Core method for efficiently optimizing multiple experimental factors (e.g., drug compound concentrations, reaction conditions) without requiring complex statistical expertise [12]. |
| Computational Complexity Theory | A framework from computer science for classifying algorithms based on their resource consumption (time, space) as a function of input size [54]. | Provides the theoretical foundation (P vs. EXP) for predicting the scalability and feasibility of computational models and data analysis pipelines in research. |
| Algorithm Visualization Tools (e.g., VisuAlgo) | Software that provides graphical representations of algorithm execution, illustrating how algorithms process data step-by-step [55] [56]. | Invaluable for educating research teams, debugging custom analysis scripts, and intuitively understanding why a particular procedure is bottlenecked. |
| Python/R Data Analysis Libraries | Programming libraries (e.g., Matplotlib, Seaborn, ggplot2) for creating custom data visualizations and performing statistical analysis [57]. | Used to implement the experimental protocol, plot runtime vs. input size, and empirically classify the complexity of in-house developed algorithms. |
This technical support guide provides a comparative analysis of two powerful experimental optimization methodologies: Evolutionary Operation (EVOP) and the Sequential Simplex Method. Within the broader thesis on optimizing multiple experimental factors, understanding the distinction and appropriate application of these techniques is paramount for efficient research and development, particularly in fields like drug development where process efficiency and continuous improvement are critical.
Evolutionary Operation (EVOP) is a philosophy for process optimization where small, deliberate changes are made to a process during routine production, without interrupting operations or producing unsatisfactory outcomes [58] [59]. Developed by George Box in the 1950s, its core principle is the continuous, real-time improvement of a process by plant operatives themselves [58]. In contrast, the Sequential Simplex Method (a specific type of EVOP) is a straightforward algorithmic procedure used to navigate multiple experimental factors to rapidly find optimal conditions [60] [61]. It is a "self-directed optimization" that works by iteratively moving away from unfavorable conditions [58].
The fundamental relationship is that Sequential Simplex is one of several design approaches that can be used to implement the broader EVOP philosophy; other approaches include full or fractional factorial designs [58].
Methodologies and Experimental Protocols
Detailed Protocol: Implementing a Sequential Simplex Experiment
The Sequential Simplex method is an efficient experimental design strategy that gives improved response after only a few experiments without complex statistical analysis [37]. The following workflow outlines the core iterative process.
Workflow Steps:
- Define the Objective and Select Factors: Clearly state the goal (e.g., maximize yield, minimize impurities) and identify the continuous process variables (factors) to be optimized [59].
- Create the Initial Simplex: A simplex is a geometric figure with one more vertex than the number of factors. For two factors, it is a triangle; for three, a tetrahedron. The initial
k+1vertexes represent the initial set of experiments [60]. - Run Experiments and Rank Responses: Perform the experiments defined by the simplex vertexes and measure the response for each. Rank the vertexes from best (B) to worst (W) response [60].
- Calculate a New Vertex: Generate a new vertex (R) by reflecting the worst vertex (W) through the centroid (P) of the remaining vertexes. The formula for the reflection is:
R = P + (P - W)[60]. - Test the New Vertex: Based on the response at R, the algorithm adapts using a set of logical rules [60]:
- If the response at R is better than the next-best (N) vertex, compute an expansion vertex (E) to move further in that promising direction.
- If the response at R is worse than N but better than W, perform a contraction (Cr) to refine the search.
- If the response at R is worse than W, perform a contraction away from this poor direction (Cw).
- Iterate Until Convergence: Replace the worst vertex with the new successful vertex, forming a new simplex. Repeat the process until the simplex converges around the optimum point or oscillates within a region of satisfactory performance [60].
Detailed Protocol: Implementing a Factorial EVOP Experiment
For comparison, here is a protocol for a traditional factorial EVOP approach, which is more aligned with the original EVOP philosophy of engaging process operators in continuous improvement [58].
Workflow Steps:
- Planning: Define the goal and identify 2-3 major process variables (factors). The changes to these factors must be small enough to not risk product quality during routine production [58] [59].
- Experimental Design: Use a simple factorial design (e.g., a 2^2 factorial with a center point) to structure the experiments. This design is run over several production cycles or phases [58].
- Execution and Data Collection: The process is run at the conditions specified by the design, and data on the critical quality attributes is collected. This is done by process operatives during normal production [58].
- Analysis: Analyze the data using basic statistical methods (e.g., calculating main effects and interaction effects) to determine the direction of improvement [58] [59].
- Adjustment and Replication: If the analysis shows a statistically significant improvement, the process operating conditions are reset to the new, better conditions. The experiment is then repeated in a new cycle to confirm the improvement or guide further optimization [59].
Comparative Analysis: Sequential Simplex vs. Factorial EVOP
The table below provides a structured, quantitative comparison of the two methods to guide method selection.
| Feature | Sequential Simplex Method | Factorial EVOP Approach |
|---|---|---|
| Core Principle | Self-directed algorithmic optimization [37] | Real-time, continuous process improvement [58] |
| Typical Number of Factors | Efficient for several factors (e.g., 3-8) [37] | Best for 2-3 key factors [59] |
| Initial Experiment Count | k + 1 (e.g., 4 expts for 3 factors) [60] | 2^k + center points (e.g., 5 expts for 2 factors) [58] |
| Information Model | Uses a simplistic geometric model of the response surface [60] | Typically uses a first-order or first-order with interaction statistical model [58] |
| Primary Goal | Rapidly find the optimum combination of factor levels [37] | Continuously and systematically move the process toward a more optimal state [58] |
| Best Application Context | Off-line R&D or process tuning; systems with short experiment times [37] [61] | On-line, high-volume production; processes subject to drift over time [58] [61] |
Frequently Asked Questions (FAQs)
1. When should I use the Sequential Simplex method over a traditional factorial EVOP?
Choose Sequential Simplex when your primary goal is speed and efficiency in optimizing a system with several continuous factors in an off-line or R&D setting [37] [61]. It is particularly useful when experiment time is short, allowing for rapid iteration. Choose a factorial EVOP approach when you need to optimize a process during routine production without interrupting output, especially for managing 2-3 critical factors and fostering a culture of continuous improvement among operators [58] [59].
2. A common issue is that my simplex seems to be circling an area but not converging on a precise optimum. What should I do?
This behavior often indicates that the simplex is navigating a region with a relatively flat response surface or that the step size is too large. This is addressed by the built-in variable-size simplex rules. The algorithm should eventually trigger a contraction (Cr or Cw), which reduces the simplex size and allows for finer exploration of the region [60]. If oscillation persists, you may be near the optimum and can average the coordinates of the best vertexes from recent cycles as your final answer.
3. My initial screening experiment did not identify a factor as significant, but I suspect it is important. Could EVOP still be useful?
Yes, this is a key strength of the optimization-first strategy. The classical approach to R&D can miss important factors during screening if their effects are non-linear or exist only in a different region of the factor space [37]. By using an efficient method like Sequential Simplex to first find a better-performing region, you can then use classical designs to model and understand the system in that new region, which may reveal the importance of the previously missed factor [37].
4. How do I ensure that my EVOP program does not produce out-of-specification product?
This is a fundamental tenet of EVOP. The changes made to the process factors must be kept within a "working region" that is known to produce acceptable, saleable product. The perturbations are small and constrained by the process's control plan limits [61]. The large amount of data collected compensates for these small changes, allowing the signal from the effect of the factor changes to be detected over the background noise of process variation [58].
The Scientist's Toolkit: Essential Reagent Solutions
The following table details key computational and statistical resources essential for conducting these optimization experiments.
| Tool or Resource | Function in Optimization | Example/Note |
|---|---|---|
| Two-Level Factorial Designs | Serves as the experimental structure for traditional EVOP; used to screen for important factors and estimate main effects [58]. | A 2^3 design to assess the impact of Temperature, Pressure, and Concentration. |
| Sequential Simplex Algorithm | The logical engine for self-directed optimization; dictates the calculation of the next experiment based on previous results [60]. | Rules for reflection, expansion, and contraction to navigate the factor space. |
| Plackett-Burman Designs | A specific, highly efficient type of fractional factorial design used for screening a large number of factors with a minimal number of runs [58] [37]. | Useful in the initial stages of the "classical" approach to identify vital few factors. |
| Central Composite Design (CCD) | A classical response surface design used to fit a full second-order model, typically after screening or after locating the region of the optimum [37]. | Used for detailed modeling and characterization of the optimum region. |
| Statistical Software/Calculator | To perform the calculations for EVOP (e.g., effects, significance) or to implement the simplex algorithm and visualize progress [59] [60]. | Can range from specialized software to simple spreadsheets programmed with the logic. |
Optimizing processes with multiple experimental factors is a core challenge in research and development. This guide compares three key methodologies: the Sequential Simplex Method, Response Surface Methodology (RSM), and Bayesian Optimization (BO). Understanding their principles, strengths, and ideal applications helps you select the best approach and troubleshoot common experimental issues.
Frequently Asked Questions (FAQs)
1. How do I choose between a model-agnostic method like Simplex and a model-based method like RSM or BO? Your choice depends on your prior knowledge of the system:
- Use Simplex when the system is poorly understood or highly complex, and you cannot assume a specific model. It relies on geometric principles to navigate the experimental space [23].
- Use RSM or BO when you have some prior knowledge and can leverage a model. RSM assumes a polynomial (often quadratic) relationship, while BO uses probabilistic models like Gaussian Processes to approximate the system's behavior [23].
2. We started with a Simplex optimization, but our progress has stalled. What should we do? This is a common issue where the simplex begins to "circle" or oscillate around a potential optimum [9]. This often indicates you are near a peak or in a noisy region.
- Solution: Apply the standard rule: reject the worst vertex and reflect it. If the new vertex is again the worst, do not reflect back. Instead, reject the second-worst vertex and reflect it to change direction [9]. If oscillations continue, consider reducing the size of your simplex to refine the search in the current region.
3. Our RSM model shows a significant "Lack of Fit." What does this mean and how can we address it? A significant Lack of Fit (LoF) means your chosen model (e.g., a quadratic polynomial) is insufficient to capture the true relationship between your factors and the response [62].
- Troubleshooting Steps:
- Check for Higher-Order Terms: Your system may require a more complex model. You might need to upgrade from a quadratic to a cubic model, if the design allows it [63].
- Consider Transformations: Explore transformations of your response variable (e.g., log, square root) to better meet model assumptions.
- Expand the Experimental Region: The optimal point might lie outside the region you investigated. Consider a follow-up experiment using the method of steepest ascent to move to a more promising region before building a new RSM model [64].
4. Bayesian Optimization is praised for its efficiency, but can it handle categorical factors, which are common in biological experiments? Yes, this is a key advantage of modern BO. Unlike traditional RSM, which is primarily designed for continuous and discrete numerical factors, BO can be adapted to handle complex design spaces that include categorical variables (e.g., different types of carbon sources or cell lines) [65]. This is achieved through the use of specialized kernels in the Gaussian Process model.
5. Is it true that Bayesian Optimization always requires fewer experiments than RSM? While BO is often more efficient, this is not an absolute rule. One study on alkaline wood delignification found that both RSM and BO found comparable optimal conditions, with BO not reducing the number of experiments but providing a more accurate model near the optimum [66]. However, other studies, particularly in complex biological systems, show BO can reduce the experimental burden by 3 to 30 times compared to statistical Design of Experiments (DoE), which includes RSM [65]. The efficiency gain depends on the problem's complexity and noise.
Comparative Analysis Tables
The following tables summarize the core characteristics, advantages, and disadvantages of each optimization method.
| Feature | Sequential Simplex | Response Surface Methodology (RSM) | Bayesian Optimization (BO) |
|---|---|---|---|
| Core Principle | Geometric operations (reflection, expansion, contraction) to navigate the factor space [9]. | Fitting a predefined polynomial model (e.g., quadratic) to the experimental data [62]. | Using a probabilistic surrogate model (e.g., Gaussian Process) and an acquisition function to guide experiments [67] [65]. |
| Model Dependency | Model-agnostic [23]. | Model-based (pre-specified model) [23]. | Model-based (adaptive, probabilistic model) [23]. |
| Experimental Strategy | Sequential | Typically parallel (full design executed before analysis) [65]. | Sequential (iterative) [65]. |
| Key Strength | Simple, intuitive, requires no assumed model. | Provides a clear, interpretable empirical model of the process. | Highly efficient for complex, noisy, or expensive-to-evaluate "black-box" functions. |
| Common Weakness | Can get stuck in local optima; may be inefficient in high-dimensional spaces [9]. | Requires many experiments upfront; model may be an oversimplification [67]. | Computational complexity; less interpretable than RSM. |
Table 2: Experimental Performance in Case Studies
| Case Study | Sequential Simplex | RSM | Bayesian Optimization | Key Finding |
|---|---|---|---|---|
| Plant-Based Protein Extrusion [67] | Not Tested | 15 trials required. Prediction error up to 61.0%. | Converged in 10-11 trials. Prediction error ≤24.5%. | BO showed superior predictive accuracy and efficiency with fewer experiments. |
| Cell Culture Media Development [65] | Not Tested | Used as a state-of-art benchmark. | Achieved improved performance with 3-30 times fewer experiments than DoE/RSM. | BO's efficiency advantage magnified with increasing factors and categorical variables. |
| Alkaline Wood Delignification [66] | Not Tested | Comparable pilot-scale results to BO. | Comparable pilot-scale results to RSM; did not reduce experiment count. | Both methods found a good optimum; BO provided a more accurate model near the optimum. |
Experimental Protocols
Protocol 1: Running a Sequential Simplex Optimization
This protocol outlines the steps for a basic (fixed-size) simplex method for two factors [9].
- Initial Simplex: Start with an initial simplex of (f+1) points (e.g., a triangle for two factors). The size and location are chosen by the experimenter.
- Run Experiments: Conduct experiments at each vertex of the simplex and measure the response.
- Rank Vertices: Rank the vertices from Best (B), Next-best (N), to Worst (W).
- Reflect: Calculate the reflection of the worst point (W) through the centroid (P) of the remaining face (B and N). The new point R is given by ( R = P + (P - W) ).
- Iterate: Run an experiment at the new point R.
- If R is better than B, consider an expansion step.
- If R is worse than N but better than W, perform a contraction.
- If R is the new worst point, perform a contraction on the W side or shrink the entire simplex.
- Terminate: Continue iterating until the simplex tightly circles an optimum or a predefined number of experiments is reached.
Protocol 2: Setting Up a Bayesian Optimization
This protocol describes the iterative workflow for BO [67] [65].
- Define the Problem: Specify the objective function (e.g., yield, purity), the factors to optimize (which can be continuous or categorical), and any constraints.
- Initial Design: Perform a small initial set of experiments (e.g., via a space-filling design like Latin Hypercube) to gather baseline data.
- Model Training: Use the collected data to train a probabilistic surrogate model, typically a Gaussian Process (GP). The GP provides a prediction and an uncertainty estimate for any point in the factor space.
- Propose Next Experiment: An acquisition function (e.g., Expected Improvement) uses the GP model to balance exploration (probing uncertain regions) and exploitation (refining good regions) to propose the most promising factor combination for the next experiment.
- Run Experiment & Update: Run the experiment at the proposed conditions, measure the result, and add this new data point to the dataset.
- Iterate: Repeat steps 3-5 until convergence (e.g., no significant improvement over several iterations) or the experimental budget is exhausted.
Workflow Visualization
The following diagram illustrates the core logical workflow for each of the three optimization methods, highlighting their fundamental differences in approach.
The Scientist's Toolkit: Key Reagents and Materials
Table 3: Essential Materials for Featured Experiments
| Item | Function/Application | Example from Literature |
|---|---|---|
| Soy Protein Concentrate (SPC) | Primary protein source providing structure and amino acid profile in plant-based meat analogues [67]. | Used with wheat gluten in high-moisture extrusion optimized by BO and RSM [67]. |
| Wheat Gluten | Forms a viscoelastic network that enhances structural integrity and fiber stability in meat analogues [67]. | Combined with SPC to create a fibrous texture in extrusion experiments [67]. |
| Commercial Cell Culture Media | Base nutrient source providing essential components for cell growth and maintenance (e.g., DMEM, RPMI) [65]. | Blended and optimized using BO to maintain PBMC (Peripheral Blood Mononuclear Cell) viability [65]. |
| Cytokines/Chemokines | Signaling proteins used to modulate cell behavior, viability, and phenotypic distribution in culture [65]. | Optimized as supplements in cell culture media using BO to maintain specific lymphocytic populations [65]. |
| Anaerobic Digestion Substrate | The biomass material (e.g., agricultural waste) that is broken down by microbes to produce biogas [68]. | Hydrothermally pretreated biomass was used as a substrate for methane production optimization with AutoML and BO [68]. |
In research utilizing the sequential simplex method for optimizing multiple experimental factors, establishing a robust validation framework is paramount. The sequential simplex is an efficient multivariate optimization algorithm that guides experimentation by moving a geometric figure (a "simplex") through the experimental parameter space to rapidly locate optimal conditions [48]. However, finding these optimal conditions is only the first step; researchers must then build confidence in these results through systematic validation and replication. This process ensures that identified optima are not merely artifacts of experimental noise or specific contextual factors but represent reliable, reproducible conditions suitable for further development and application.
The core challenge lies in distinguishing between local optima—favorable conditions specific to a particular experimental setup—and globally optimal conditions that hold across different contexts. This is where structured validation, particularly through replication strategies, becomes essential. By implementing a framework that incorporates both within-study and across-study replication [69], researchers can progressively build evidence supporting the reliability and generalizability of their optimized conditions.
Core Validation Framework: The V3 Principles
Adapted from the Digital Medicine Society's (DiMe) framework and tailored for experimental optimization, the V3 framework provides a comprehensive structure for validation [70]. This approach segments the validation process into three distinct but interconnected phases:
Verification: Ensuring that the fundamental experimental components—instruments, sensors, and data acquisition systems—accurately capture and store raw data. In simplex optimization, this involves verifying that parameter controls (e.g., temperature, flow rate, concentration) truly reflect their set points and that measurement systems provide accurate readings across the experimental range.
Analytical Validation: Assessing the precision and accuracy of the algorithms and processes that transform raw data into meaningful experimental outcomes. For sequential simplex, this includes validating that the algorithm correctly interprets responses, appropriately calculates new vertex points, and accurately terminates at the true optimum rather than being misled by experimental noise.
Clinical (or Contextual) Validation: Confirming that the optimized conditions meaningfully reflect the desired biological, chemical, or physical states relevant to their context of use [70]. In pharmaceutical development, this means demonstrating that factors optimized using the simplex method (e.g., reaction conditions, formulation parameters) actually produce the desired therapeutic outcomes, not just statistical improvements.
Troubleshooting Guides & FAQs
Common Optimization Challenges and Solutions
| Challenge | Root Cause | Solution Approach | Validation Step |
|---|---|---|---|
| Apparent Convergence at Suboptimal Conditions | Local optimum trapping; insufficient step size; noisy response measurements | Implement robustness testing by restarting from different initial simplex points; adjust reflection/expansion coefficients [48] | Across-study replication with modified initial conditions [69] |
| High Variability in Replicated Optima | Poorly controlled experimental parameters; highly stochastic systems; insufficient response measurement precision | Increase experimental controls; implement replication at each simplex point; use weighted averaging of responses [48] | Verification of measurement systems; analytical validation of response functions [70] |
| Failure to Reproduce Optimized Conditions | Unaccounted-for parameter interactions; uncontrolled environmental factors; instrument calibration drift | Conduct full parameter interaction analysis; control environmental variables; implement regular calibration protocols | Independent replication by different researchers [69]; verification of experimental conditions [70] |
| Algorithm Cycling or Early Termination | Improper convergence criteria; degeneracy in simplex structure; numerical precision issues | Implement Bland's rule for pivot selection [71]; adjust convergence thresholds; increase numerical precision in calculations | Analytical validation of algorithm implementation; verification with benchmark problems |
| Discrepancy Between Optimized Conditions and Final Outcomes | Inappropriate response function; missing critical parameters; scale-up effects | Re-evaluate response function relevance; include additional potentially significant factors; implement staged optimization | Contextual validation across different scales [70]; replication with extension [69] |
Frequently Asked Questions
Q1: How many replication studies are typically needed to have confidence in optimized conditions identified via simplex methods?
The number of required replications depends on the consequences of failure and system variability. For high-stakes applications (e.g., pharmaceutical formulation), a minimum of 3-5 successful independent replications under varying conditions provides reasonable confidence [69]. The progression should include:
- Direct replication: Reproducing the exact conditions to verify initial results
- Replication with extension: Testing under modified but relevant conditions (different operators, equipment, or environmental conditions)
- Independent replication: Conducted by different research teams to eliminate potential biases [69]
Q2: What specific steps should I take when my replication fails to reproduce previously optimized conditions?
- Verify measurement systems: Recalibrate all instruments and verify parameter controls [70]
- Audit experimental conditions: Document and compare all environmental factors, reagent sources, and equipment settings
- Convergence analysis: Verify that both the original and replication studies achieved true convergence by examining simplex behavior near the optimum [48]
- Parameter sensitivity analysis: Determine if small variations in critical parameters could explain the discrepancy
- Progressive complexity testing: Return to simpler systems to isolate the source of discrepancy
Q3: How can I distinguish between algorithmic failures and genuine experimental variability when validation fails?
Implement a triangulation approach:
- Benchmark testing: Apply your simplex implementation to well-characterized benchmark problems with known optima [48]
- Cross-validation with alternative methods: Compare results with other optimization approaches (e.g., DoE, OVAT) for the same system [48]
- Algorithm verification: Implement diagnostic tracking of simplex operations (vertex calculations, reflection decisions, convergence checks) [71]
- Experimental controls: Include standardized reference points throughout the optimization process to separate algorithmic performance from experimental variability
Q4: What documentation is essential for facilitating successful replication of simplex optimization studies?
- Initial simplex configuration: Complete specification of starting vertices, including all parameter values
- Algorithm parameters: Reflection, expansion, contraction, and shrinkage coefficients; convergence criteria [48]
- Response function definition: Complete mathematical definition of how experimental outcomes are transformed into the response guiding the simplex
- Environmental context: Full documentation of laboratory conditions, equipment models, reagent sources, and personnel involvement
- Raw data logging: Complete records of all experimental runs, including failed attempts and outliers
- Decision trail: Documentation of all simplex moves, including the reasoning behind each step when deviations from standard practice occur
Experimental Protocols for Validation
Protocol for Direct Replication Studies
Purpose: To verify that optimized conditions identified in an initial simplex optimization produce equivalent results when reproduced under identical conditions.
Materials:
- Same equipment models and calibration standards as original study
- Identical reagent sources and lot numbers
- Equivalent environmental controls (temperature, humidity, lighting)
- Standardized measurement instruments with recent calibration certificates
Procedure:
- Pre-replication audit: Verify that all conditions match the original study's documentation
- Calibration verification: Confirm that all parameter controls (temperature, flow rates, concentrations) accurately reflect set points [70]
- Baseline testing: Reproduce 2-3 key data points from the original study's trajectory to verify comparable system behavior
- Full replication: Execute the complete set of optimized conditions identified in the original study
- Response measurement: Collect response data using identical measurement protocols and timing
- Statistical comparison: Apply equivalence testing with predetermined equivalence bounds (e.g., 5% difference in primary response)
Acceptance Criteria: Response measurements must fall within pre-specified equivalence bounds of the original results for all critical response variables.
Protocol for Replication with Extension
Purpose: To assess the robustness and generalizability of optimized conditions under modified but relevant circumstances [69].
Materials:
- Different equipment models with similar specifications
- Alternative reagent sources or lot numbers
- Modified environmental conditions within operational ranges
- Additional operators with varying experience levels
Procedure:
- Parameter modification matrix: Systematically identify which parameters will be varied and to what extent
- Staged implementation: Begin with minimal changes, progressively increasing modifications
- Response monitoring: Track both primary responses and additional stability indicators
- Interaction analysis: Document any parameter-interaction effects on optimal conditions
- Boundary mapping: Identify the operational boundaries beyond which optimized conditions fail
Analysis: Response surface methodology around the identified optimum can help characterize the robustness of the optimized conditions.
Research Reagent Solutions & Essential Materials
| Reagent/Material | Function in Optimization | Validation Considerations |
|---|---|---|
| Reference Standards (e.g., USP standards, certified reference materials) | Provides benchmark for measurement verification and cross-experiment comparison [70] | Certificate of analysis traceability; stability monitoring; proper storage conditions |
| Calibration Solutions (e.g., pH buffers, concentration standards) | Ensures measurement accuracy throughout optimization process [70] | Fresh preparation or expiration monitoring; verification against certified standards |
| Process Solvents/Reagents (multiple lots from different suppliers) | Tests robustness of optimized conditions to material variations [69] | Documentation of source, lot number, and impurity profiles; pre-use testing |
| Stability Indicators (e.g., internal standards, degradation markers) | Monitors system stability during extended optimization and replication studies | Demonstrated selectivity and sensitivity; stability under experimental conditions |
| System Suitability Test Materials | Verifies overall system performance before critical replication attempts [70] | Well-characterized response profile; established acceptance criteria |
Workflow Visualization
Sequential Simplex Validation Workflow
Replication Strategy Decision Framework
Implementation Checklist for Researchers
Pre-Optimization Phase
- Document all equipment specifications and calibration dates
- Establish baseline measurement precision through repeated controls
- Define explicit convergence criteria for the simplex algorithm
- Specify primary and secondary response functions
- Plan replication strategy concurrently with optimization design
During Optimization
- Log all experimental runs, including failed attempts
- Monitor system stability through control measurements
- Document all simplex moves and decision points
- Periodically verify measurement system calibration
- Capture environmental conditions and potential perturbations
Post-Optimization Validation
- Execute direct replication under identical conditions
- Conduct replication with systematically varied parameters
- Arrange independent replication by separate research team
- Perform statistical equivalence testing against original results
- Document any discrepancies and investigate root causes
- Update optimization parameters if boundary limitations are identified
This comprehensive validation framework ensures that optimal conditions identified through sequential simplex optimization are not merely statistical artifacts but represent robust, reproducible configurations suitable for further development and application across the pharmaceutical and chemical sciences.
Frequently Asked Questions (FAQs)
Q1: The simplex method works well in my initial experiments with a few factors, but performance degrades dramatically as I add more. Is this expected behavior? Yes, this is a well-documented phenomenon. The simplex algorithm operates by moving along the edges of a geometric polytope defined by your constraints. In low-dimensional factor spaces, this polytope has relatively few extreme points. However, as dimensionality increases, the number of vertices grows exponentially, a challenge often called the "curse of dimensionality." The algorithm may need to traverse a significantly longer path to find the optimum, drastically increasing computation time [1] [72].
Q2: My high-dimensional experimental optimization problem seems impossible to solve with the standard sequential simplex. Are there specific conditions that make such problems tractable? Successful high-dimensional optimization almost always relies on sparsity. This is the principle that, despite the large number of potential factors, only a small subset (low effective sparsity, k) truly influences the response. When this condition is met, and you have a sufficient sample size (n), specialized methods like the Lasso (Least Absolute Shrinkage and Selection Operator) can be applied to identify the influential factors before optimization, making the subsequent simplex search highly efficient [73].
Q3: What does "degeneracy" mean in the context of the simplex method, and why does it occur more frequently in high-dimensional spaces? Degeneracy occurs when more constraints than necessary intersect at a single extreme point. When this happens, the simplex method may perform "pivot" operations that change the set of active constraints without moving to a new point in the factor space. In the worst case, this can lead to cycling, where the algorithm loops indefinitely between the same bases. In high-dimensional spaces, the complex geometry makes such overlapping constraints more probable, increasing the risk of degeneracy and stalled progress [72].
Q4: For high-dimensional factor screening, should I abandon the simplex method entirely in favor of newer interior-point methods? Not necessarily. While interior-point methods can be efficient for dense, high-dimensional problems, the simplex method and its variants remain state-of-the-art for many applications. A key advantage is re-optimization. If you are solving a series of related problems—for instance, slightly adjusting your experimental constraints—the simplex method can use the previous solution as a "warm start," often converging much faster than starting from scratch. The dual simplex method is particularly powerful for this [72].
Troubleshooting Guides
Problem 1: Slow Convergence in High-Dimensional Factor Space
Symptoms: The optimization process takes impractically long to find an optimum after adding more experimental factors.
| Potential Cause | Diagnostic Steps | Solution |
|---|---|---|
| Exponential Growth of Search Space | Check the ratio of your experimental runs (n) to the number of factors (p). A very small n/p ratio is a strong indicator. | 1. Factor Screening: Use a preliminary screening design (e.g., Plackett-Burman) to identify the most influential factors. 2. Regularization: Apply techniques like Lasso regression to enforce sparsity, effectively reducing the active dimensions [73]. |
| Poor Initial Starting Point | Observe if the algorithm spends a long time in a region with poor performance. | Employ a Phase I simplex method to find a feasible starting point that is closer to the optimal region before beginning the main optimization (Phase II) [1] [72]. |
Problem 2: Algorithm Failure or Infeasible Solution
Symptoms: The solver returns an "infeasible" error or fails to converge, even when a feasible solution is believed to exist.
| Potential Cause | Diagnostic Steps | Solution |
|---|---|---|
| Incorrect Problem Formulation | Validate that all constraints are correctly specified and that no conflicting requirements exist. | 1. Re-formulate the problem in standard form by converting inequalities to equalities using slack and surplus variables [1]. 2. Use the Phase I simplex method to systematically find a feasible solution or confirm true infeasibility. |
| Numerical Instability in High Dimensions | Check for very large or very small numerical coefficients in the constraint matrix, which can cause rounding errors. | Scale the columns and rows of your constraint matrix to improve its numerical condition. This is especially critical when p is large. |
Experimental Protocols for Dimensionality Analysis
Protocol 1: Benchmarking Simplex Performance Across Dimensionality
Objective: To quantitatively assess the performance of the sequential simplex method as the number of experimental factors increases.
Materials:
- Computational software with a simplex solver (e.g., R, Python with SciPy, MATLAB).
- Standardized test functions for optimization (e.g., quadratic functions with known minima).
Methodology:
- Problem Generation: For dimensions p = 2, 5, 10, 20, 50, and 100, generate a random linear programming problem in standard form: Maximize ( \mathbf{c}^T\mathbf{x} ), subject to ( A\mathbf{x} \leq \mathbf{b} ) and ( \mathbf{x} \geq 0 ) [1].
- Execution: For each p, run the simplex algorithm 10 times with different random seeds and record the average number of:
- Pivot operations required.
- Iterations until convergence.
- Computation time in seconds.
- Data Analysis: Plot the recorded metrics against the dimension p. The resulting curves will visually demonstrate the computational cost of increasing dimensionality.
Protocol 2: Evaluating the Impact of Sparsity
Objective: To demonstrate how the sparsity of influential factors affects the solvability of high-dimensional optimization problems.
Methodology:
- Setup: Fix a high dimension (e.g., p = 100) and a sample size (e.g., n = 50), creating a "large p, small n" scenario [73].
- Sparsity Variation: Systematically vary the effective sparsity k (the number of non-zero, influential factors) from 2 to 20.
- Application of LASSO: For each k value, generate data where only k factors have a real effect. Use the Lasso to select the important factors.
- Optimization: Run the simplex method on the reduced factor set identified by the Lasso.
- Success Metric: Record the success rate in finding the true global optimum for each k. This will generate data for a phase diagram showing the transition from solvable to unsolvable regions based on the n/p and k/n ratios [73].
Workflow and Relationship Visualizations
Diagram 1: Optimization Path Based on Dimensionality
Diagram 2: Simplex Performance in Different Dimensionality
Research Reagent Solutions
Table: Key Computational and Methodological Tools
| Item Name | Function/Brief Explanation | Application Context |
|---|---|---|
| Standard Form Converter | Transforms linear inequalities into equalities by adding slack and surplus variables, a prerequisite for the simplex algorithm [1]. | Essential pre-processing step for all linear programs before applying the simplex method. |
| Phase I Simplex Method | A auxiliary linear program used to find an initial basic feasible solution (extreme point) or prove that none exists (infeasibility) [1] [72]. | Used when a obvious starting point for the main optimization (Phase II) is not available. |
| Dual Simplex Method | A variant that maintains optimality while working towards feasibility, ideal for re-optimization after modifying constraints [72]. | Highly efficient for solving a series of closely related problems, common in iterative experimental design. |
| Lasso (L1 Regularization) | A regression-based method that performs variable selection and regularization by penalizing the absolute size of coefficients, enforcing sparsity [73]. | Crucial for factor screening in high-dimensional "large p, small n" problems to identify the few active factors. |
| Solver with Column Generation | An advanced implementation that only considers variables (columns) that can improve the objective, avoiding full problem enumeration [72]. | Used for problems with a vast number of potential variables, such as in complex scheduling or logistics. |
Conclusion
The sequential simplex method remains a robust, efficient optimization technique particularly well-suited for biomedical researchers and drug development professionals working with complex, multi-factor experimental systems. Its model-agnostic nature provides distinct advantages when system behavior is poorly understood or highly complex, while its geometric approach efficiently handles factor interactions that confound one-variable-at-a-time methodologies. Recent theoretical advances have strengthened the mathematical foundation of simplex-based methods, addressing long-standing concerns about worst-case performance while explaining their consistent practical efficiency. For the future of biomedical research, sequential simplex offers particular promise in optimizing bioprocess development, analytical method validation, and drug formulation design—areas where traditional optimization approaches often prove inadequate. As experimental complexity continues to increase, the integration of simplex methods with modern computational approaches and automated experimentation platforms will likely expand their utility in accelerating discovery and development timelines across the pharmaceutical and biotechnology sectors.
References
- 1. Simplex algorithm [https://en.wikipedia.org/wiki/Simplex_algorithm]
- 2. George Dantzig [https://en.wikipedia.org/wiki/George_Dantzig]
- 3. Use Statistics to Experiment, Surface New Data and Boost ... [https://www.forbes.com/sites/rkoselka/2025/11/20/how-to-experiment-with-more-real-data-to-boost-performance/]
- 4. 7 Multi-Factor Experiments – Modern Experimental Design [https://www.refsmmat.com/courses/727/lecture-notes/multifactor.html]
- 5. The Simplex Method [https://personal.utdallas.edu/~scniu/OPRE-6201/documents/LP4-Simplex.html]
- 6. Experimental design optimization [https://mbb-team.github.io/VBA-toolbox/wiki/Optimizing-the-experimental-design/]
- 7. Linear Programming Optimization: The Simplex Method [https://towardsdatascience.com/linear-programming-optimization-the-simplex-method-b2f912e4c6fd/]
- 8. Dimensional Simplex - an overview [https://www.sciencedirect.com/topics/mathematics/dimensional-simplex]
- 9. Simplex Algorithm - an overview [https://www.sciencedirect.com/topics/mathematics/simplex-algorithm]
- 10. Geometric Algorithms for Neural Combinatorial ... [https://arxiv.org/html/2510.24039v1]
- 11. Sequential Simplex Method [https://link.springer.com/rwe/10.1007/978-3-030-54621-2_598-1]
- 12. Simplex optimization: A tutorial approach and recent ... [https://www.sciencedirect.com/science/article/abs/pii/S0026265X1500168X]
- 13. Nelder–Mead method [https://en.wikipedia.org/wiki/Nelder%E2%80%93Mead_method]
- 14. Breaking down the Nelder Mead algorithm [https://brandewinder.com/2022/03/31/breaking-down-Nelder-Mead/]
- 15. Visualizing Nelder-Mead Optimization | Alex Dowad Computes [https://alexdowad.github.io/visualizing-nelder-mead/]
- 16. Nelder-Mead method - OxRSE Training - University of Oxford [https://train.rse.ox.ac.uk/material/HPCu/scientific_computing/optimisation/06-nelder-mead]
- 17. Simplex Method - an overview [https://www.sciencedirect.com/topics/earth-and-planetary-sciences/simplex-method]
- 18. The hybrid experimental simplex algorithm – An alternative ... [https://www.sciencedirect.com/science/article/pii/S0003267012009592]
- 19. Example of a Simplex Lattice Design [https://www.jmp.com/support/help/en/19.0/jmp/example-of-a-simplex-lattice-design.shtml]
- 20. 5.5.4.2. Simplex-lattice designs [https://www.itl.nist.gov/div898/handbook/pri/section5/pri542.htm]
- 21. Simplex Designs: Example [https://help.reliasoft.com/weibull/content/simplex_designs__example.htm]
- 22. The Use of Factorial Design and Simplex Optimization to ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC7411898/]
- 23. The Evolution of Optimization Methods | by Victor Guiller [https://medium.com/@victor.guiller/the-evolution-of-optimization-methods-e7616e97343f]
- 24. Efficient Optimization With Ax, an Open Platform for Adaptive ... [https://engineering.fb.com/2025/11/18/open-source/efficient-optimization-ax-open-platform-adaptive-experimentation/]
- 25. Protein Expression in E. coli: A Powerful System ... [https://www.sinobiological.com/resource/protein-review/ecoli-protein-expression-system]
- 26. Promiscuous protein biotinylation by Escherichia coli biotin ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC2286582/]
- 27. Recombinant Protein Production - Part 1 - 2025 Archive [https://www.chi-peptalk.com/25/protein-expression-production]
- 28. A Highâ€Throughput Multi Well Plate–Based Approach for ... [https://pmc.ncbi.nlm.nih.gov/articles/PMC12590519/]
- 29. The statistical simplex method for experimental ... [https://www.sciencedirect.com/science/article/pii/S1570794605801270]
- 30. Are You Sure You Understand USP ? [https://www.chromatographyonline.com/view/are-you-sure-you-understand-usp-621-]
- 31. Employment of Sequential Simplex Method of Optimization ... [https://jopcr.com/articles/employment-of-sequential-simplex-method-of-optimization-in-the-development-of-fast-dissolving-tablets-of-clozapine]
- 32. Chromatography [https://www.usp.org/harmonization-standards/pdg/excipients/chromatography]
- 33. troubleshooting! [http://www.chromforum.org/viewtopic.php?t=6036]
- 34. Design and evaluation of an improved simplex method [https://www.sciencedirect.com/science/article/abs/pii/S000326709800316X]
- 35. Optimisation under capsis [https://capsis.cirad.fr/capsis/documentation/optimisation]
- 36. Experimental Design Approaches in Method Optimization [https://www.chromatographyonline.com/view/experimental-design-approaches-method-optimization-0]
- 37. Optimization - PMC - NIH [https://pmc.ncbi.nlm.nih.gov/articles/PMC6644965/]
- 38. Challenges in measuring and understanding biological noise [https://pmc.ncbi.nlm.nih.gov/articles/PMC7611518/]
- 39. Making use of noise in biological systems [https://www.sciencedirect.com/science/article/abs/pii/S0079610723000020]
- 40. Noise-guided tuning of synthetic protein waves in living cells [https://pmc.ncbi.nlm.nih.gov/articles/PMC11957142/]
- 41. Noise in Biology - PMC - PubMed Central - NIH [https://pmc.ncbi.nlm.nih.gov/articles/PMC4033672/]
- 42. How thoughtful experimental design can empower ... [https://www.nature.com/articles/s41467-025-62616-x]
- 43. The Relationship Between Biological Noise and Its ... [https://www.mdpi.com/2079-7737/14/4/349]
- 44. Improving SNR and Determining Optimal Excitation Levels [https://www.ni.com/en/support/documentation/supplemental/21/making-accurate-strain-measurements-improving-snr-and-determining-optimal-excitation-levels.html?srsltid=AfmBOop8BMMA4oin2KPIq4W0eqCOTfEnkCHpjJ3H59b8smrZo9CbS9eH]
- 45. Methods of Multi-Criteria Optimization of Technological ... [https://www.mdpi.com/2227-7390/12/18/2856]
- 46. A comparison of Evolutionary Operation and Simplex for ... [https://www.sciencedirect.com/science/article/abs/pii/S0169743914002007]
- 47. Simple Trick to Dramatically Improve Speed of Convergence [https://www.datasciencecentral.com/simple-trick-to-dramatically-improve-speed-of-convergence/]
- 48. Self-optimising processes and real-time ... - RSC Publishing [https://pubs.rsc.org/en/content/articlehtml/2020/re/d0re00081g]
- 49. Beyond Smoothed Analysis: Analyzing the Simplex Method ... [https://arxiv.org/html/2510.21613v1]
- 50. Simplex Method [https://neos-guide.org/guide/algorithms/simplex/]
- 51. Hybrid-LP: Finding advanced starting points for simplex, ... [https://www.sciencedirect.com/science/article/abs/pii/S0305054810001322]
- 52. A Hybrid Global Optimization Algorithm Involving Simplex ... [https://link.springer.com/chapter/10.1007/3-540-45718-6_73]
- 53. Difference Between Exponential and Polynomial ... [https://www.geeksforgeeks.org/dsa/difference-between-exponential-and-polynomial-complexities/]
- 54. Time complexity [https://en.wikipedia.org/wiki/Time_complexity]
- 55. VisuAlgo: visualising data structures and algorithms through ... [https://visualgo.net/]
- 56. Algorithm Visualization Tools [https://www.meegle.com/en_us/topics/algorithm/algorithm-visualization-tools]
- 57. Data Visualization: Tools - Research Guides [https://research.lib.buffalo.edu/dataviz/tools]
- 58. Tools in Waiting: Time for Evolutionary Operation [https://www.pharmtech.com/view/tools-waiting-time-evolutionary-operation]
- 59. Evolutionary Operation (EVOP) [https://www.benchmarksixsigma.com/forum/topic/39446-evolutionary-operation-evop/]
- 60. EVOPtimizer [http://www.engineeredsoftware.com/sso.asp]
- 61. (PDF) EVOP Design of Experiments [https://www.academia.edu/72726016/EVOP_Design_of_Experiments]
- 62. Response Surface Methodology (RSM) in Design of ... [https://www.6sigma.us/six-sigma-in-focus/response-surface-methodology-rsm/]
- 63. v25.0 » Tutorials » One-Factor RSM [https://www.statease.com/docs/v25.0/tutorials/one-factor-rsm/]
- 64. Lesson 11: Response Surface Methods and Designs [https://online.stat.psu.edu/stat503/lesson/11]
- 65. Accelerating cell culture media development using ... [https://www.nature.com/articles/s41467-025-61113-5]
- 66. Traditional or adaptive design of experiments? A pilot ... [https://www.sciencedirect.com/science/article/pii/S2405844024005152]
- 67. Plant-based protein extrusion optimization - PubMed Central [https://pmc.ncbi.nlm.nih.gov/articles/PMC12336687/]
- 68. Accelerating integrated prediction, analysis and targeted ... [https://www.sciencedirect.com/science/article/abs/pii/S1364032124004143]
- 69. Replication in Scientific Validation and Identification of ... [https://asatonline.org/for-parents/becoming-a-savvy-consumer/role-of-replication-in-scientific-validation/]
- 70. Validation framework for in vivo digital measures - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC11751004/]
- 71. The Simplex Method [https://labs.acme.byu.edu/Volume2/Simplex/Simplex.html]
- 72. Primal and Dual Simplex Methods - Science4All [http://www.science4all.org/article/simplex-methods/]
- 73. Statistical challenges of high-dimensional data - PMC [https://pmc.ncbi.nlm.nih.gov/articles/PMC2865881/]